Tidy datasets are all alike, but every messy dataset is messy in its own way.
–Hadley Wickham (cf. Leo Tolstoy)
A great deal of data both does and should live in tabular formats; to put it flatly, this means formats that have rows and columns. In a theoretical sense, it is possible to represent every collection of structured data in terms of multiple "flat" or "tabular" collections if we also have a concept of relations. Relational database management systems (RDBMS) have had a great deal of success since 1970, and a very large part of all the world's data lives in RDBMS's. Another large share lives in formats that are not relational as such, but that are nonetheless tabular, wherein relationships may be imputed in an ad hoc, but uncumbersome, way.
As the Preface mentioned, the data ingestion chapters will concern themselves chiefly with structural or mechanical problems that make data dirty. Later in the book we will focus more on content or numerical issues in data.
This chapter discusses tabular formats including CSV, spreadsheets, SQL databases, and scientific array storage formats. The last sections look at some general concepts around data frames, which will typically be how data scientists manipulate tabular data. Much of this chapter is concerned with the actual mechanics of ingesting and working with a variety of data formats, using several different tools and programming languages. The Preface discusses why I wish to remain language agnostic—or multilingual—in my choices. Where each format is prone to particular kinds of data integrity problems, special attention is drawn to that. Actually remediating those characteristic problems is largely left until later chapters; detecting them is the focus of our attention here.
As The Hitchhiker's Guide to the Galaxy is humorously inscribed: "Don't Panic!" We will explain in much more detail the concepts mentioned here.
We run the setup code that will be standard throughout this book. As the Preface mentions, each chapter can be
run in
full, assuming available configuration files have been utilized. The prompts you see throughout this book, such
as
In [1]: and
practice in Python to use import *, we do so here to bring in many names without a long block of imports.
from src.setup import *
%load_ext rpy2.ipython
%%capture --no-stdout err %%R library(tidyverse)
With our various Python and R libraries now available, let us utilize them to start cleaning data.
After every war someone has to tidy up.
–Maria Wisława Anna Szymborska
Concepts:
Hadley Wickham and Garrett Grolemund, in their excellent and freely available book, [R for Data Science](https://r4ds.had.co.nz/), promote the concept of "tidy data." The Tidyverse collection of R packages attempt to realize this concept in concrete libraries. Wickham and Grolemund's idea of tidy data has a very close intellectual forebearer in the concept of database normalization, which is a large topic addressed in depth neither by them nor in this current book. The canonical reference on database normalization is C. J. Date's An Introduction to Database Systems (Addison Wesley; 1975 and numerous subsequent editions).
In brief, tidy data carefully separates variables (the columns of a table; also called features or fields) from observations (the rows of a table; also called samples). At the intersection of these two, we find values, one data item (datum) in each cell. Unfortunately, the data we encounter is often not arranged in this useful way, and it requires normalization. In particular, what are really values are often represented either as columns or as rows instead. To demonstrate what this means, let us consider an example.
Returning to the small elementary school class we presented in the Preface, we might encounter data looking like this:
students = pd.read_csv('data/students-scores.csv')
students
| Last Name | First Name | 4th Grade | 5th Grade | 6th Grade | |
|---|---|---|---|---|---|
| 0 | Johnson | Mia | A | B+ | A- |
| 1 | Lopez | Liam | B | B | A+ |
| 2 | Lee | Isabella | C | C- | B- |
| 3 | Fisher | Mason | B | B- | C+ |
| 4 | Gupta | Olivia | B | A+ | A |
| 5 | Robinson | Sophia | A+ | B- | A |
This view of the data is easy for humans to read. We can see trends in the scores each student received over several years of education. Moreover, this format might lend itself to useful visualizations fairly easily.
# Generic conversion of letter grades to numbers
def num_score(x):
to_num = {'A+': 4.3, 'A': 4, 'A-': 3.7,
'B+': 3.3, 'B': 3, 'B-': 2.7,
'C+': 2.3, 'C': 2, 'C-': 1.7}
return x.map(lambda x: to_num.get(x, x))
This next cell uses a "fluent" programming style that may look unfamiliar to some Python programmers. I discuss this style in the section below on data frames. The fluent style is used in many data science tools and languages. For example, this is typical Pandas code that plots the students' scores by year.
(students
.set_index('Last Name')
.drop('First Name', axis=1)
.apply(num_score)
.T
.plot(title="Student score by year")
.legend(bbox_to_anchor=(1, .75))
)
plt.savefig("img/(Ch01)Student score by year");
This data layout exposes its limitations once the class advances to 7th grade, or if we were to obtain 3rd grade information. To accommodate such additional data, we would need to change the number and position of columns, not simply add additional rows. It is natural to make new observations or identify new samples (rows), but usually awkward to change the underlying variables (columns).
The particular class level (e.g. 4th grade) that a letter grade pertains to is, at heart, a value not a variable. Another way to think of this is in terms of independent variables versus dependent variables. Or in machine learning terms, features versus target. In some way, the class level might correlate with or influence the resulting letter grade; perhaps the teachers at the different levels have different biases, or children of a certain age lose or gain interest in schoolwork, for example.
For most analytic purposes, this data would be more useful if we make it tidy (normalized) before further
processing.
In Pandas, the DataFrame.melt() method can perform this tidying. We pin some of the columns as
id_vars, and we set a name for the combined columns as a variable and the letter grade as a single
new
column. This Pandas method is slightly magical, and takes some practice to get used to. The key thing is that it
preserves data, simply moving it between column labels and data values.
students.melt(
id_vars=["Last Name", "First Name"],
var_name="Level",
value_name="Score"
).set_index(['First Name', 'Last Name', 'Level'])
| Score | |||
|---|---|---|---|
| First Name | Last Name | Level | |
| Mia | Johnson | 4th Grade | A |
| Liam | Lopez | 4th Grade | B |
| Isabella | Lee | 4th Grade | C |
| Mason | Fisher | 4th Grade | B |
| ... | ... | ... | ... |
| Isabella | Lee | 6th Grade | B- |
| Mason | Fisher | 6th Grade | C+ |
| Olivia | Gupta | 6th Grade | A |
| Sophia | Robinson | 6th Grade | A |
18 rows × 1 columns
In the R Tidyverse, the procedure is similar. Do not worry about the extra Jupyter magic
%%capture, this
simply quiets extra informational messages sent to STDERR that would distract from the printed book. A
tibble that we see here is simply a kind of data frame that is preferred in the Tidyverse.
%%capture --no-stdout err
%%R
library('tidyverse')
studentsR <- read_csv('data/students-scores.csv')
studentsR
── Column specification ────────────────────────────────────────────────────────────────── cols( `Last Name` = col_character(), `First Name` = col_character(), `4th Grade` = col_character(), `5th Grade` = col_character(), `6th Grade` = col_character() ) # A tibble: 6 x 5 `Last Name` `First Name` `4th Grade` `5th Grade` `6th Grade` <chr> <chr> <chr> <chr> <chr> 1 Johnson Mia A B+ A- 2 Lopez Liam B B A+ 3 Lee Isabella C C- B- 4 Fisher Mason B B- C+ 5 Gupta Olivia B A+ A 6 Robinson Sophia A+ B- A
Within the Tidyverse, specifically within the tidyr package, there is a function
pivot_longer() that is similar to Pandas' .melt(). The aggregation names and values
have
parameters spelled names_to and values_to, but the operation is the same.
%%capture --no-stdout err
%%R
studentsR <- read_csv('data/students-scores.csv')
studentsR %>%
pivot_longer(c(`4th Grade`, `5th Grade`, `6th Grade`),
names_to = "Level",
values_to = "Score")
── Column specification ────────────────────────────────────────────────────────────────── cols( `Last Name` = col_character(), `First Name` = col_character(), `4th Grade` = col_character(), `5th Grade` = col_character(), `6th Grade` = col_character() ) # A tibble: 18 x 4 `Last Name` `First Name` Level Score <chr> <chr> <chr> <chr> 1 Johnson Mia 4th Grade A 2 Johnson Mia 5th Grade B+ 3 Johnson Mia 6th Grade A- 4 Lopez Liam 4th Grade B 5 Lopez Liam 5th Grade B 6 Lopez Liam 6th Grade A+ 7 Lee Isabella 4th Grade C 8 Lee Isabella 5th Grade C- 9 Lee Isabella 6th Grade B- 10 Fisher Mason 4th Grade B 11 Fisher Mason 5th Grade B- 12 Fisher Mason 6th Grade C+ 13 Gupta Olivia 4th Grade B 14 Gupta Olivia 5th Grade A+ 15 Gupta Olivia 6th Grade A 16 Robinson Sophia 4th Grade A+ 17 Robinson Sophia 5th Grade B- 18 Robinson Sophia 6th Grade A
The simple example above gives you a first feel for tidying tabular data. To reverse the tidying operation that
moves
variables (columns) to values (rows), the pivot_wider() function in tidyr can be used. In Pandas
there
are several related methods on DataFrames, including .pivot(), .pivot_table(), and
.groupby() combined with .unstack(), which can create columns from rows (and do many
other
things too).
Having looked at the idea of tidyness as a general goal for tabular, let us being looking at specific data formats, beginning with comma-separated values and fixed-width files.
Speech sounds cannot be understood, delimited, classified and explained except in the light of the tasks which they perform in language.
–Roman Jakobson
Concepts:
Delimited text files, especially comma-separated values (CSV) files, are ubiquitous. These are text files that put multiple values on each line, and separate those values with some semi-reserved character, such as a comma. They are almost always the exchange format used to transport data between other tabular representations, but a great deal of data both starts and ends life as CSV, perhaps never passing through other formats.
Reading delimited files is not the fastest way of reading data from disk into RAM memory, but it is also not the slowest. Of course, that concern only matters for large-ish data sets, not for the small data sets that make up most of our work as data scientists (small nowadays means roughly "fewer then 100k rows").
There are a great number of deficits in CSV files, but also some notable strengths. CSV files are the format second most susceptible to structural problems. All formats are generally equally prone to content problems, which are not tied to the format itself. Spreadsheets like Excel are, of course, by a very large margin the worst format for every kind of data integrity concern.
At the same time, delimited formats—or fixed-width text formats—are also almost the only ones you can easily open and make sense of in a text editor or easily manipulate using command-line tools for text processing. Thereby delimited files are pretty much the only ones you can fix fully manually without specialized readers and libraries. Of course, formats that rigorously enforce structural constraints do avoid some of the need to do this. Later in this chapter, and in the next two chapters, a number of formats that enforce structure more are discussed.
One issue that you could encounter in reading CSV or other textual files is the actual character set encoding may not be the one you expect, or that is the default on your current system. In this age of Unicode, this concern is diminishing, but only slowly, and archival files continue to exist. This topic is discussed in chapter 3 (Data Ingestion – Repurposing Data Sources) in the section Custom Text Formats.
As a quick example, suppose you have just received a medium sized CSV file, and you want to make a quick sanity check on it. At this stage, we are concerned about whether the file is formatted correctly at all. We can do this with command-line tools, even if most libraries are likely to choke on them (such as shown in the next code cell). Of course, we could also use Python, R, or another general-purpose language if we just consider the lines as text initially.
# Use try/except to avoid full traceback in example
try:
pd.read_csv('data/big-random.csv')
except Exception as err:
print_err(err)
ParserError Error tokenizing data. C error: Expected 6 fields in line 75, saw 8
What went wrong there? Let us check.
%%bash # What is the general size/shape of this file? wc data/big-random.csv
100000 100000 4335846 data/big-random.csv
Great! 100,000 rows; but there is some sort of problem on line 75 according to Pandas (and perhaps on other lines as well). Using a single piped bash command which counts commas per line might provide insight. We could absolutely perform this same analysis in Python, R, or other languages; however, being familiar with command-line tools is a benefit to data scientists in performing one-off analyses like this.
%%bash
cat data/big-random.csv |
tr -d -c ',\n' |
awk '{ print length; }' |
sort |
uniq -c
46 3
99909 5
45 7
So we have figured out already that 99,909 of the lines have the expected 5 commas. But 46 have a deficit and 45 a surplus. Perhaps we will simply discard the bad lines, but that is not altogether too many to consider fixing by hand, even in a text editor. We need to make a judgement, on a per problem basis, about both the relative effort and reliability of automation of fixes versus manual approaches. Let us take a look at a few of the problem rows.
%%bash
grep -C1 -nP '^([^,]+,){7}' data/big-random.csv | head
74-squarcerai,45,quiescenze,12,scuoieremo,70 75:fantasmagorici,28,immischiavate,44,schiavizzammo,97,sfilzarono,49 76-interagiste,50,repentagli,72,attendato,95 -- 712-resettando,58,strisciato,46,insaldai,62 713:aspirasse,15,imbozzimatrici,70,incanalante,93,succhieremo,41 714-saccarometriche,18,stremaste,12,hindi,19 -- 8096-squincio,16,biascicona,93,solisti,70 8097:rinegoziante,50,circoncidiamo,83,stringavate,79,stipularono,34
Looking at these lists of Italian words and integers of slightly varying number of fields does not immediately illuminate the nature of the problem. We likely need more domain or problem knowledge. However, given that fewer than 1% of rows are a problem, perhaps we should simply discard them for now. If you do decide to make a modification such as removing rows, then versioning the data, with accompanying documentation of change history and reasons, becomes crucial to good data and process provenance.
This next cell uses a regular expression to filter the lines in the "almost CSV" file. The pattern may appear
confusing, but regular expressions provide a compact way of describing patterns in text. The match in
pat
indicates that from beginning of a line (^) until the end of that line ($) there are
exactly
five repetitions of character sequences that do not include commas, each followed by one comma
([^,]+,).
import re
pat = re.compile(r'^([^,]+,){5}[^,]*$')
with open('data/big-random.csv') as fh:
lines = [l.strip().split(',')
for l in fh if re.match(pat, l)]
pd.DataFrame(lines)
| 0 | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| 0 | infilaste | 21 | esemplava | 15 | stabaccavo | 73 |
| 1 | abbadaste | 50 | enartrosi | 85 | iella | 54 |
| 2 | frustulo | 77 | temporale | 83 | scoppianti | 91 |
| 3 | gavocciolo | 84 | postelegrafiche | 93 | inglesizzanti | 63 |
| ... | ... | ... | ... | ... | ... | ... |
| 99905 | notareschi | 60 | paganico | 64 | esecutavamo | 20 |
| 99906 | rispranghiamo | 11 | schioccano | 44 | imbozzarono | 80 |
| 99907 | compone | 85 | disfronderebbe | 19 | vaporizzavo | 54 |
| 99908 | ritardata | 29 | scordare | 43 | appuntirebbe | 24 |
99909 rows × 6 columns
In the code we managed, within Python, to read all rows without formatting problems. We could also have used
the
pd.read_csv() parameter error_bad_lines=False to achieve the same effect, but walking
through it in plain Python and Bash gives you a better picture of why they are excluded.
Let us return to some virtues and deficits of CSV files. Here when we mention CSV, we really mean any kind of
delimited file. And specifically, text files that store tabular data nearly always use a single character for a
delimiter, and end rows/records with a newline (or carriage return and newline in legacy formats). Other than
commas,
probably the most common delimiters you will encounter are tabs and the pipe character |. However,
nearly
all tools are more than happy to use an arbitrary character.
Fixed-width files are similar to delimited ones. Technically they are different in that, although they are line oriented, they put each field of data in specific character positions within each line. An example is used in the next code cell below. Decades ago, when Fortran and Cobol were more popular, fixed-width formats were more prevalent; my perception is that their use has diminished in favor of delimited files. In any case, fixed-width textual data files have most of the same pitfalls and strengths as do delimited ones.
The Bad
Columns in delimited or flat files do not carry a data type, being simply text values. Many tools will (optionally) make guesses about the data type, but these are subject to pitfalls. Moreover, even where the tools accurately guess the broad type category (i.e. string vs. integer vs. real number) they cannot guess the specific bit length desired, where that matters.
Likewise, the representation used for "real" numbers is not encoded—most systems deal with IEEE-754 floating-point numbers of some length, but occasionally decimals of some specific length are more appropriate for a purpose.
The most typical way that type inference goes wrong is where the initial records in some data set have an apparent pattern, but later records deviate from this. The software library may infer one data type but later encounter strings that cannot be cast as such. "Earlier" and "later" here can have several different meanings.
For out-of-core data frame libraries like Vaex and Dask (Python libraries) that read lazily, type heuristics might be applied to a first few records (and perhaps some other sampling) but will not see those strings that do not follow the assumed pattern. However, later might also mean months later, when new data arrives.partnum
Most data frame libraries are greedy about inferring data types—although all will allow manual specification to shortcut inference.
For many layouts, data frame libraries can guess a fixed-width format and infer column positions and data types
(where it cannot guess, we could manually specify). But the guesses about data types can go wrong. For example,
viewing the raw text, we see a fixed-width layout in parts.fwf.
%%bash cat data/parts.fwf
Part_No Description Maker Price (USD) 12345 Wankle rotary engine Acme Corporation 555.55 67890 Sousaphone Marching Inc. 333.33 2468 Feather Duster Sweeps Bros 22.22 A9922 Area 51 metal fragment No Such Agency 9999.99
Reading in a few rows of data:
Reading this with Pandas correctly infers the intended column positions for the fields.
df = pd.read_fwf('data/parts.fwf', nrows=3)
df
| Part_No | Description | Maker | Price (USD) | |
|---|---|---|---|---|
| 0 | 12345 | Wankle rotary engine | Acme Corporation | 555.55 |
| 1 | 67890 | Sousaphone | Marching Inc. | 333.33 |
| 2 | 2468 | Feather Duster | Sweeps Bros | 22.22 |
df.dtypes
Part_No int64 Description object Maker object Price (USD) float64 dtype: object
We deliberately only read the start of the parts.fwf file. From those first few rows, Pandas made
a type
inference of int64 for the Part_No column.
Let us read the entire file. Pandas does the "right thing" here: Part_No becomes a generic object,
i.e.
string. However, if we had a million rows instead, and the heuristics Pandas uses, for speed and memory
efficiency,
happened to limit inference to the first 100,000 rows, we might not be so lucky.
df = pd.read_fwf('data/parts.fwf')
df
| Part_No | Description | Maker | Price (USD) | |
|---|---|---|---|---|
| 0 | 12345 | Wankle rotary engine | Acme Corporation | 555.55 |
| 1 | 67890 | Sousaphone | Marching Inc. | 333.33 |
| 2 | 2468 | Feather Duster | Sweeps Bros | 22.22 |
| 3 | A9922 | Area 51 metal fragment | No Such Agency | 9999.99 |
df.dtypes # type of `Part_No` changed
Part_No object Description object Maker object Price (USD) float64 dtype: object
R tibbles behave the same as Pandas, with the minor difference that data type imputation always uses 1000 rows (and will discard values if inconsistencies occur thereafter). Pandas can be configured to read all rows for inference, but by default reads a dynamically determined number. Pandas will sample more rows than R does, but still only approximately tens of thousands. The R collections data.frame and data.table are likewise similar. Let us reading the same file as above using R.
%%capture --no-stdout err
%%R
read_table('data/parts.fwf')
── Column specification ────────────────────────────────────────────────────────────────── cols( Part_No = col_character(), Description = col_character(), Maker = col_character(), `Price (USD)` = col_double() ) # A tibble: 4 x 4 Part_No Description Maker `Price (USD)` <chr> <chr> <chr> <dbl> 1 12345 Wankle rotary engine Acme Corporation 556. 2 67890 Sousaphone Marching Inc. 333. 3 2468 Feather Duster Sweeps Bros 22.2 4 A9922 Area 51 metal fragment No Such Agency 10000.
Again, the first three rows are consistent with an integer data type, although this is inaccurate for later rows.
%%R
read_table('data/parts.fwf',
n_max = 3,
col_types = cols("i", "-", "f", "n"))
# A tibble: 3 x 3
Part_No Maker `Price (USD)`
<int> <fct> <dbl>
1 12345 Acme Corporation 556.
2 67890 Marching Inc. 333.
3 2468 Sweeps Bros 22.2
Delimited files—but not so much fixed-width files—are prone to escaping issues. In particular, CSVs specifically often contain descriptive fields that sometimes contain commas within the value itself. When done right, this comma should be escaped. It is often not done right in practice.
CSV is actually a family of different dialects, mostly varying in their escaping conventions. Sometimes spacing before or after commas is treated differently across dialects as well. One approach to escaping is to put quotes around either every string value, or every value of any kind, or perhaps only those values that contain the prohibited comma. This varies by tool and by the version of the tool. Of course, if you quote fields, there is potentially a need to escape those quotes; usually this is done by placing a backslash before the quote character when it is part of the value.
An alternate approach is to place a backslash before those commas that are not intended as a delimeter but
rather
part of a string value (or numeric value that might be formatted, e.g. $1,234.56). Guessing the
variant
can be a mess, and even single files are not necessarily self consistent between rows, in practice (often
different
tools or versions of tools have touched the data).
Tab-separated and pipe-separated formats are often chosen with the hope of avoiding escaping issues. This works
to an
extent. Both tabs and pipe symbols are far less common in ordinary prose. But both still wind up occurring in
text
occasionally, and all the escaping issues come back. Moreover, in the face of escaping, the simplest tools
sometimes
fail. For example, the bash command cut -d, will not work in these cases, nor will Python's
str.split(','). A more custom parser becomes necessary, albeit a simple one compared to
full-fledged
grammars. Python's standard library csv module is one such custom parser.
The corresponding danger for fixed-width files, in contrast to delimited ones, is that values become too long. Within a certain line position range you can have any codepoints whatsoever (other than newlines). But once the description or name that someone thought would never be longer than, say, 20 characters becomes 21 characters, the format fails.
A special consideration arises around reading datetime formats. Data frame libraries that read datetime values typically have an optional switch to parse certain columns as datetime formats. Libraries such as Pandas support heuristic guessing of datetime formats; the problem here is that applying a heuristic to each of millions of rows can be exceedingly slow. Where a date format is uniform, using a manual format specifier can make it several orders of magnitude faster to read. Of course, where the format varies, heuristics are practically magic; and perhaps we should simply marvel that the dog can talk at all rather than criticize its grammar. Let us look at a Pandas' attempt to guess datetimes for each row of a tab-separated file.
%%bash # Notice many date formats cat data/parts.tsv
Part_No Description Date Price (USD) 12345 Wankle rotary 2020-04-12T15:53:21 555.55 67890 Sousaphone April 12, 2020 333.33 2468 Feather Duster 4/12/2020 22.22 A9922 Area 51 metal 04/12/20 9999.99
# Let Pandas make guesses for each row
# VERY SLOW for large tables
parts = pd.read_csv('data/parts.tsv',
sep='\t', parse_dates=['Date'])
parts
| Part_No | Description | Date | Price (USD) | |
|---|---|---|---|---|
| 0 | 12345 | Wankle rotary | 2020-04-12 15:53:21 | 555.55 |
| 1 | 67890 | Sousaphone | 2020-04-12 00:00:00 | 333.33 |
| 2 | 2468 | Feather Duster | 2020-04-12 00:00:00 | 22.22 |
| 3 | A9922 | Area 51 metal | 2020-04-12 00:00:00 | 9999.99 |
We can verify that the dates are genuinely a datetime data type within the DataFrame.
parts.dtypes
Part_No object Description object Date datetime64[ns] Price (USD) float64 dtype: object
We have looked at some challenges and limitations of delimited and fixed-width formats, let us consider their considerable advantages as well.
The Good
The biggest strength of CSV files, and their delimited or fixed-width cousins, is the ubiquity of tools to read
and
write them. Every library dealing with data frames or arrays, across every programming language, knows how to
handle
them. Most of the time the libraries parse the quirky cases pretty well. Every spreadsheet program imports and
exports
as CSV. Every RDBMS—and most non-relational databases as well—imports and exports as CSV. Most programmers' text
editors even have facilities to make editing CSV easier. Python has a standard library module csv
that
processes many dialects of CSV (or other delimited formats) as a line-by-line record reader.
The fact that so very many structurally flawed CSV files live in the wild shows that not every tool
handles
them entirely correctly. In part, that is probably because the format is simple enough to almost work
without
custom tools at all. I have myself, in a "throw-away script," written
print(",".join([1,2,3,4]), file=csv) countless times; that works well, until it doesn't. Of course,
throw-away scripts become fixed standard procedures for data flow far too often.
The lack of type specification is often a strength rather than a weakness. For example, the part numbers mentioned a few pages ago may have started out always being integers as an actual business intention, but later on a need arose to use non-integer "numbers." With formats that have a formal type specifier, we generally have to perform a migration and copy to move old data into a new format that follows the loosened or revised constraints.
One particular case where a data type change happens especially often, in my experience, is with finite-width character fields. Initially some field is specified as needing 5, or 15, or 100 characters for its maximum length, but then a need for a longer string is encountered later, and a fixed table structure or SQL database needs to be modified to accommodate the longer length. Even more often—especially with databases—the requirement is underdocumented, and we wind up with a data set filled with truncated strings that are of little utility (and perhaps permanently lost data).
Text formats in general are usually flexible in this regard. Delimited files—but not fixed-width files—will happily contain fields of arbitrary length. This is similarly true of JSON data, YAML dataconfig, XML data, log files, and some other formats that simply utilize text, often with line-oriented records. In all of this, data typing is very loose and only genuinely exists in the data processing steps. That is often a great virtue.
A related "looseness" of CSV and similar formats is that we often indefinitely aggregate multiple CSV files that follow the same informal schema. Writing a different CSV file for each day, or each hour, or each month, of some ongoing data collection is very commonplace. Many tools, such as Dask and Spark will seamlessly treat collections of CSV files (matching a glob pattern on the file system) as a single data set. Of course, in tools that do not directly support this, manual concatenation is still not difficult. But under the model of having a directory that contains an indefinite number of related CSV snapshots, presenting it as a single common object is helpful.
The libraries that handle families of CSV files seamlessly are generally lazy and distributed. That is, with these tools, you do not typically read in all the CSV files at once, or at least not into the main memory of a single machine. Rather, various cores or various nodes in a cluster will each obtain file handles to individual files, and the schema information will be inferred from only one or a few of the files, with actual processing deferred until a specific (parallel) computation is launched. Splitting processing of an individual CSV file across cores is not easily tractable since a reader can only determine where a new record begins by scanning until it finds a newline.
While details of the specific APIs of libraries for distributed data frames is outside the scope of this book, the fact that parallelism is easily possible given an initial division of data into many files is a significant strength for CSV as a format. Dask in particular works by creating many Pandas DataFrames and coordinating computation upon all of them (or those needed for a given result) with an API that exactly copies the same methods of individual Pandas objects.
# Generated data files with random values
from glob import glob
# Use glob() function to identify files matching pattern
glob('data/multicsv/2000-*.csv')[:8] # ... and more
['data/multicsv/2000-01-27.csv', 'data/multicsv/2000-01-26.csv', 'data/multicsv/2000-01-06.csv', 'data/multicsv/2000-01-20.csv', 'data/multicsv/2000-01-13.csv', 'data/multicsv/2000-01-22.csv', 'data/multicsv/2000-01-21.csv', 'data/multicsv/2000-01-24.csv']
We read this family of CSV files into one virtualized DataFrame that acts like a Pandas DataFrame, even if loading it with Pandas would require more memory than our local system allows. In this specific example, the collection of CSV files is not genuinely too large for a modern workstation to read into memory; but when it becomes so, using some distributed or out-of-core system like Dask is necessary to proceed at all.
import dask.dataframe as dd
df = dd.read_csv('data/multicsv/2000-*-*.csv',
parse_dates=['timestamp'])
print("Total rows:", len(df))
df.head()
Total rows: 2592000
| timestamp | id | name | x | y | |
|---|---|---|---|---|---|
| 0 | 2000-01-01 00:00:00 | 979 | Zelda | 0.802163 | 0.166619 |
| 1 | 2000-01-01 00:00:01 | 1019 | Ingrid | -0.349999 | 0.704687 |
| 2 | 2000-01-01 00:00:02 | 1007 | Hannah | -0.169853 | -0.050842 |
| 3 | 2000-01-01 00:00:03 | 1034 | Ursula | 0.868090 | -0.190783 |
| 4 | 2000-01-01 00:00:04 | 1024 | Ingrid | 0.083798 | 0.109101 |
When we require some summary to be computed, Dask will coordinate workers to aggregate on each individual DataFrame, then aggregate those aggregations. There are more nuanced issues of what operations can be reframed in this "map-reduce" style and which cannot, but that is the general idea (and the Dask or Spark developers have thought about this for you so you do not have to).
df.mean().compute()
id 999.965606 x 0.000096 y 0.000081 dtype: float64
Having looked at some pros and cons of working with CSV data, let us turn to another format where a great deal of data is stored. Unfortunately, for spreadsheets, there are almost exclusively cons.
Drugs are bad. m'kay. You shouldn't do drugs. m'kay. If you do them you're bad, because drugs are bad. m'kay. It's a bad thing to do drugs, so don't be bad by doing drugs. m'kay.
–Mr. Mackay (South Park)
Concepts:
Edward Tufte, that brilliant doyen of information visualization, wrote an essay called The Cognitive Style of Powerpoint: Pitching Out Corrupts Within. Amongst his observations is that the manner in which slide presentations, and Powerpoint specifically, hides important information more than it reveals it, was a major or even main cause of the 2003 Columbia space shuttle disaster. Powerpoint is anathema to clear presentation of information.
To no less of a degree, spreadsheets in general, and Excel in particular, are anathema to effective data science. While perhaps not as much as in CSV files, a great share of the world's data lives in Excel spreadsheets. There are numerous kinds of data corruption that are the special realm of spreadsheets. As a bonus, data science tools read spreadsheets much more slowly than they do every other format, while spreadsheets also have hard limits on the amount of data they can contain that other formats do not impose.
Most of what spreadsheets do to make themselves convenient for their users makes them bad for scientfic reproducibility, data science, statistics, data analysis, and related areas.computation Spreadsheets have apparent rows and columns in them, but nothing enforces consistent use of those, even within a single sheet. Some particular feature often lives in column F for some rows, but the equivalent thing is in column H for other rows, for example. Contrast this with a CSV file or an SQL table; for these latter formats, while all the data in a column is not necessarily good data, it generally must pertain to the same feature.
Another danger of spreadsheets is not around data ingestion, per se, at all. Computation within spreadsheets is spread among many cells in no obvious or easily inspectable order, leading to numerous large-scale disasterous consequences (loss of billions in financial transaction; a worldwide economic planning debacle; a massive failure of Covid-19 contact tracing in the UK). The European Spreadsheet Risks Interest Group is an entire organization devoted to chronicling such errors. They present a number of lovely quotes, including this one:
There is a literature on denial, which focuses on illness and the fact that many people with terminal illnesses deny the seriousness of their condition or the need to take action. Apparently, what is very difficult and unpleasant to do is difficult to contemplate. Although denial has only been studied extensively in the medical literature, it is likely to appear whenever required actions are difficult or onerous. Given the effortful nature of spreadsheet testing, developers may be victims of denial, which may manifest itself in the form of overconfidence in accuracy so that extensive testing will not be needed. –Ray Panko, 2003
In procedural programming (including object-oriented programming), actions flow sequentially through code, with clear locations for branches or function calls; even in functional paradigms, compositions are explicitly stated. In spreadsheets it is anyone's guess what computation depends on what else, and what data ranges are actually included. Errors can occasionally be found accidentally, but program analysis and debugging are *nearly* impossible. Users who know only, or mostly, spreadsheets will likely object that *some* tools exist to identify dependencies within a spreadsheet; this is technically true in the same sense as that many goods transported by freight train could also be carried on a wheelbarrow.
Moreover, every cell in a spreadsheet can have a different data type. Usually the type is assigned by heuristic guesses within the spreadsheet interface. These are highly sensitive to the exact keystrokes used, the order cells are entered, whether data is copy/pasted between blocks, and numerous other things that are both hard to predict and that change between every version of every spreadsheet software program. Infamously, for example, Excel interprets the gene name 'SEPT2' (Septin 2) as a date (at least in a wide range of versions). Compounding the problem, the interfaces of spreadsheets make determining the data type for a given cell uncomfortably difficult.
Let us start with an example. The screenshot below is of a commonplace and ordinary looking spreadsheet. Yes, some values are not aligned in their cells exactly consistently, but that is purely an aesthetic issue. The first problem that jumps out at us is the fact that one sheet is being used to represent two different (in this case related) tables of data. Already this is going to be difficult to make tidy.
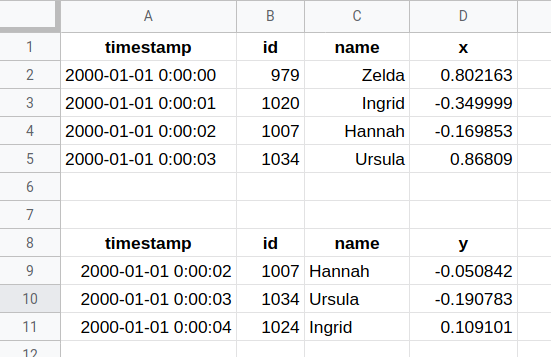
Image: Excel Pitfalls
If we simply tell Pandas (or specifically the supporting openpyxl library) to try to make sense of this file, it makes a sincere effort and applies fairly intelligent heuristics. It does not crash, to its credit. Other DataFrame libraries will be similar, with different quirks you will need to learn. But what went wrong that we can see initially?
# Default engine `xlrd` might have bug in Python 3.9
pd.read_excel('data/Excel-Pitfalls.xlsx',
sheet_name="Dask Sample", engine="openpyxl")
| timestamp | id | name | x | |
|---|---|---|---|---|
| 0 | 2000-01-01 00:00:00 | 979 | Zelda | 0.802163 |
| 1 | 2000-01-01 0:00:01 | 1019.5 | Ingrid | -0.349999 |
| 2 | 2000-01-01 00:00:02 | 1007 | Hannah | -0.169853 |
| 3 | 2000-01-01 00:00:03 | 1034 | Ursula | 0.86809 |
| 4 | timestamp | id | name | y |
| 5 | 2000-01-01 00:00:02 | 1007 | Hannah | -0.050842 |
| 6 | 2000-01-01 00:00:03 | 1034 | Ursula | -0.190783 |
| 7 | 2000-01-01 00:00:04 | 1024 | Ingrid | 0.109101 |
Right away we can notice that the id column contains a value 1019.5 that was invisible in the
spreadsheet display. Whether that column is intended as a floating-point or an integer is not obvious at this
point.
Moreover, notice that visually the date on that same row looks slightly wrong. We will come back to this.
As a first step, we can, with laborious manual intervention, pull out the two separate tables we actually care about. Pandas is actually a little bit too smart here—it will, by default, ignore the data typing actually in the spreadsheet and do inference similar to what it does with a CSV file. For this purpose, we tell it to use the data type actually stored by Excel. Pandas' inference is not a panacea, but it is a useful option at times (it can fix some, but not all, of the issues we note below; however, other things are made worse). For the next few paragraphs, we wish to see the raw data types stored in the spreadsheet itself.
df1 = pd.read_excel('data/Excel-Pitfalls.xlsx',
nrows=5, dtype=object, engine="openpyxl")
df1.loc[:2] # Just look at first few rows
| timestamp | id | name | x | |
|---|---|---|---|---|
| 0 | 2000-01-01 00:00:00 | 979 | Zelda | 0.802163 |
| 1 | 2000-01-01 00:00:01 | 1019.5 | Ingrid | -0.349999 |
| 2 | 2000-01-01 00:00:02 | 1007 | Hannah | -0.169853 |
We can read the second implicit table as well by using the pd.read_excel() parameter
skiprows.
pd.read_excel('data/Excel-Pitfalls.xlsx', skiprows=7, engine="openpyxl")
| timestamp | id | name | y | |
|---|---|---|---|---|
| 0 | 2000-01-01 00:00:02 | 1007 | Hannah | -0.050842 |
| 1 | 2000-01-01 00:00:03 | 1034 | Ursula | -0.190783 |
| 2 | 2000-01-01 00:00:04 | 1024 | Ingrid | 0.109101 |
If we look at the data types read in, we will see they are all Python objects to preserve the various cell types. But let us look more closely at what we actually have.
df1.dtypes
timestamp datetime64[ns] id object name object x object dtype: object
The timestamps in this particular small example are all reasonable to parse with Pandas. But real-life spreadsheets often provide something much more ambiguous, often impossible to parse as dates. Look above at the screenshot of a spreadsheet to notice that the data type is invisible in the spreadsheet itself. We can find the Python data type of the generic object stored in each cell.
# Look at the stored data type of each cell
tss = df1.loc[:2, 'timestamp']
for i, ts in enumerate(tss):
print(f"TS {i}: {ts}\t{ts.__class__.__name__}")
TS 0: 2000-01-01 00:00:00 Timestamp TS 1: 2000-01-01 00:00:01 Timestamp TS 2: 2000-01-01 00:00:02 Timestamp
The Pandas to_datetime() function is idempotentidempotent and would have run if
we had
not specifically disabled it in by using dtype=object in the pd.read_excel() call.
However,
many spreadsheets are far messier, and the conversion will simply not succeed, producing an object
column
in any case. Particular cells in a column might contain numbers, formulae, or strings looking nothing like dates
(or
sometimes strings looking just enough like date string that a human, but not a machine, might guess the intent;
say
'Decc 23,, 201.9').
Let us look at using pd.to_datetime().
pd.to_datetime(tss)
0 2000-01-01 00:00:00 1 2000-01-01 00:00:01 2 2000-01-01 00:00:02 Name: timestamp, dtype: datetime64[ns]
Other columns pose a similar difficulty. The values that look identical in the spreadsheet view of the
id column are actually a mixture of integers, floating-point numbers, and strings. It is
conceivable that such was the intention, but in practice it is almost always an accidental result of
the ways
that spreadsheets hide information from their users. By the time these data sets arrive on your data science
desk,
they are merely messy, and the causes are lost in the sands of time. Let us look at the data types in the
id column.
# Look at the stored data type of each cell
ids = df1.loc[:3, 'id']
for i, id_ in enumerate(ids):
print(f"id {i}: {id_}\t{id_.__class__.__name__}")
id 0: 979 int id 1: 1019.5 float id 2: 1007 int id 3: 1034 str
Of course tools like Pandas can type cast values subsequent to reading them, but we require domain-specific
knowledge
of the data set to know what cast is appropriate. Let us cast data using the .astype() method:
ids.astype(int)
0 979 1 1019 2 1007 3 1034 Name: id, dtype: int64
Putting together the cleanup we mention, we might carefully type our data in a manner similar to the following.
# Only rows through index `3` are useful
# We are casting to more specific data types
# based on domain and problem knowledge
df1 = df1.loc[0:3].astype(
{'id': np.uint16,
'name': pd.StringDtype(),
'x': float})
# datetimes require conversion function, not just type
df1['timestamp'] = pd.to_datetime(df1.timestamp)
print(df1.dtypes)
timestamp datetime64[ns] id uint16 name string x float64 dtype: object
df1.set_index('timestamp')
| id | name | x | |
|---|---|---|---|
| timestamp | |||
| 2000-01-01 00:00:00 | 979 | Zelda | 0.802163 |
| 2000-01-01 00:00:01 | 1019 | Ingrid | -0.349999 |
| 2000-01-01 00:00:02 | 1007 | Hannah | -0.169853 |
| 2000-01-01 00:00:03 | 1034 | Ursula | 0.868090 |
What makes spreadsheets harmful is not principally their underlying data formats. Non-ancient versions of Excel (.xlsx), LibreOffice (OpenDocument, .ods), and Gnumeric (.gnm) have all adopted a similar format at the byte level. That is, they all store their data in XML formats, then compress those to save space. As I mentioned, this is slower than other approaches, but that concern is secondary.
If one of these spreadsheet formats were used purely as an exchange format among structured tools, they would be perfectly suitable to preserve and represent data. It is instead the social and user interface (UI) elements of spreadsheets that make them dangerous. The "tabular" format of Excel combines the worst elements of untyped CSV and strongly typed SQL databases. Rather than assign a data type by column/feature, it allows type assignments per cell.
Per-cell typing is almost always the wrong thing to do for any data science purpose. It neither allows flexible decisions by programming tools (either using inference or type declaration APIs) nor does it enforce consistency of different values that should belong to the same feature at the time data is stored. Moreover, the relatively free-form style of entry in the user interfaces of spreadsheets does nothing to guide users away from numerous kinds of entry errors (not only data typing, but also various misalignments within the grid, accidental deletions or insertions, and so on). Metaphorically, the danger posed by spreadsheet UIs resembles the concept in tort law of an "attractive nuisance"—they do not directly create the harm, but they make harm exceedingly likely with minor inattention.
Unfortunately, there do not currently exist any general purpose data entry tools in widespread use. Database entry forms could serve the purpose of enforcing structure on data entry, but they are limited for non-programmatic data exploration. Moreover, use of structured forms, whatever the format where the data might be subsequently stored, currently requires at least a modicum of software development effort, and many general users of spreadsheets lack this ability. Something similar to a spreadsheet, but that allowed locking data type constraints on columns, would be a welcome addition to the world. Perhaps one or several of my readers will create and popularize such tools.
For now, the reality is that many users will create spreadsheets that you will need to extract data from as a data scientist. This will inevitably be more work for you than if you were provided a different format. But think carefully about the block regions and tabs/sheets that are of actual relevance, to the problem-required data types for casts, and about how to clean unprocessable values. With effort the data will enter your data pipelines.
We can turn now to well structured and carefully date typed formats; those stored in relational databases.
At the time, Nixon was normalizing relations with China. I figured that if he could normalize relations, then so could I.
–E. F. Codd (inventor of relational database theory)
Concepts:
Relational databases management systems (RDBMS) are enormously powerful and versatile. For the most part, their requirement of strict column typing and frequent use of formal foreign keys and constraints is a great boon for data science. While specific RDBMS's vary greatly in how well normalized, indexed, and designed they are—not every organization has or utilizes a database engineer specifically—even somewhat informally assembled databases have many desirable properties for data science. Not all relational databases are tidy, but they all take you several large steps in that direction.
Working with relational databases requires some knowledge of Structured Query Language (SQL). For small data, and perhaps for medium-sized data, you can get by with reading entire tables into memory as data frames. Operations like filtering, sorting, grouping, and even joins can be performed with data frame libraries. However, it is much more efficient if you are able to do these kinds of operations directly at the database level; it is an absolute necessity when working with big data. A database that has millions or billions of records, distributed across tens or hundreds of related tables, can itself quickly produce the hundreds of thousands of rows (tuples) that you need for the task at hand. But loading all of these rows into memory is either unnecessary or simply impossible.
There are many excellent books and tutorials on SQL. I do not have a specific one to recommend over others, but
finding a suitable text to get up to speed—if you are not already—is not difficult. The general concepts of
GROUP BY, JOIN, and WHERE clauses are the main things you should know as
a data
scientist. If you have a bit more control over the database you pull data from, knowing something about how to
intelligently index tables, and optimize slow queries by reformulation and looking at EXPLAIN
output, is
helpful. However, it is quite likely that you, as a data scientist, will not have full access to database
administration. If you do have such access: be careful!
For this book, I use a local PostgreSQL server to illustrate APIs. I find that PostgreSQL is
vastly
better at query optimization than is its main open source competitor, MySQL. Both behave
equally well
with careful index tuning, but generally PostgreSQL is much faster for queries that must be optimized on an ad
hoc
basis by the query planner. In general, almost all of the APIs I show will be nearly identical across drivers in
Python or in R (and in most other languages) whether you use PostgreSQL, MySQL, Oracle DB, DB2, SQL Server, or
any
other RDBMS. The Python DB-API, in particular, is well standardized across drivers. Even the single-file RDBMS,
SQLite3, which is included in the Python standard library is almost DB-API compliant (and
.sqlite is a very good storage format).
Within the setup.py module that is loaded by each chapter and is available within the source code
repository, some database setup is performed. If you run some of the functions contained therein, you will be
able to
create generally the same configuration on your system as I have on the one where I am writing this. Actual
installation of an RDBMS is not addressed in this book; see the instructions accompanying your database
software. But
a key, and simple step, is creating a connection to the DB.
# Similar with adapter other than psycopg2
con = psycopg2.connect(database=db, host=host,
user=user, password=pwd)
This connection object will be used in subsequent code in this book. We also create an engine
object
that is an SQLAlchemy wrapper around a connection that adds some enhancements. Some libraries
like
Pandas require using an engine rather than only a connection. We can create that as follows:
engine = create_engine(
f'postgresql://{user}:{pwd}@{host}:{port}/{db}')
I used the Dask data created earlier in this chapter to populate a table with the following schema. These metadata values are defined in the RDBMS itself. Within this section, we will work with the elaborate and precise data types that relational databases provide.
| Column | Data Type | Data Width |
|---|---|---|
| index | integer | 32 |
| timestamp | timestamp without time zone | None |
| id | smallint | 16 |
| name | character | 10 |
| x | numeric | 6 |
| y | real | 24 |
This is the same data structure created in the previous Dask discussion, but I have somewhat arbitrarily
imposed more
specific data types on the fields. The PostgreSQL "data width" shown is a bit odd; it mixes bit length with byte
length depending on the type. Moreover, for the floating-point y it shows the bit length of the
mantissa,
not of the entire 32-bit memory word. But in general we can see that different columns have different specific
types.
When designing tables, database engineers generally try to choose data widths that are sufficient for the purpose, but also as small as the requirement allows. If you need to store billions of person ages, for example, a 256-bit integer could certainly hold those numbers, but an 8-bit integer can also hold all the values that can occur using 1/32nd as much storage space.
Using the Python DB-API loses some data type information. It does pretty well, but Python does not
have a
full range of native types. The fractional numbers are accurately stored as either Decimal or
native
floating-point, but the specific bit lengths are lost. Likewise, the integer is a Python integer of unbounded
size.
The name strings are always 10 characters long, but for most purposes we probably want to apply
str.rstrip() (strip whitespace at right end) to take off the surrounding whitespace.
# Function connect_local() spelled out in chapter 4 (Anomaly Detection)
con, engine = connect_local()
cur = con.cursor()
cur.execute("SELECT * FROM dask_sample")
pprint(cur.fetchmany(2))
[(3456,
datetime.datetime(2000, 1, 2, 0, 57, 36),
941,
'Alice ',
Decimal('-0.612'),
-0.636485),
(3457,
datetime.datetime(2000, 1, 2, 0, 57, 37),
1004,
'Victor ',
Decimal('0.450'),
-0.68771815)]
Unfortunately, we lose even more data type information using Pandas (at least as of Pandas 1.0.1 and SQLAlchemy 1.3.13, current as of this writing). Pandas is able to use the full type system of NumPy, and even adds a few more custom types of its own. This richness is comparable to—but not necessarily identical with—the type systems provided by RDBMS's (which, in fact, vary from each other as well, especially in extension types). However, the translation layer only casts to basic string, float, int, and date types.
Let us read a PostgreSQL table into Pandas, and then examine what native data types were utilized to approximate that SQL data.
df = pd.read_sql('dask_sample', engine, index_col='index')
df.tail(3)
| timestamp | id | name | x | y | |
|---|---|---|---|---|---|
| index | |||||
| 5676 | 2000-01-02 01:34:36 | 1041 | Charlie | -0.587 | 0.206869 |
| 5677 | 2000-01-02 01:34:37 | 1017 | Ray | 0.311 | 0.256218 |
| 5678 | 2000-01-02 01:34:38 | 1036 | Yvonne | 0.409 | 0.535841 |
The specific dtypes within the DataFrame are:
df.dtypes
timestamp datetime64[ns] id int64 name object x float64 y float64 dtype: object
Although it is a bit more laborious, we can combine these techniques and still work with our data within a friendly data frame, but using more closely matched types (albeit not perfectly matched to the DB). The two drawbacks here are:
object columnsLet us endeavor to choose better data types for our data frame. We probably need to determine the precise types
from
the documentation of our RDBMS, since few people have the PostgreSQL type codes memorized. The DB-API cursor
object
has a .description attribute that contains column type codes.
cur.execute("SELECT * FROM dask_sample")
cur.description
(Column(name='index', type_code=23), Column(name='timestamp', type_code=1114), Column(name='id', type_code=21), Column(name='name', type_code=1042), Column(name='x', type_code=1700), Column(name='y', type_code=700))
We can introspect to see the Python types used in the results. Of course, these do not carry the bit lengths of
the
DB with them, so we will need to manually choose these. Datetime is straightforward enough to put into Pandas's
datetime64[ns] type.
rows = cur.fetchall() [type(v) for v in rows[0]]
[int, datetime.datetime, int, str, decimal.Decimal, float]
Working with Decimal numbers is tricker than other types. Python's standard library decimal module
complies with IBM’s General Decimal Arithmetic Specification;
unfortunately, databases do not. In particular, the IBM 1981 spec (with numerous updates) allows each
operation to be performed within some chosen "decimal context" that gives precision, rounding rules,
and
other things. This is simply different from having a decimal precision per column, with no
specific
control of rounding rules. We can usually ignore these nuances; but when they bite us, they can bite hard. The
issues
arise more in civil engineering and banking/finance than they do with data science as such, but these are
concerns to
be aware of.
In the next cell, we cast several columns to specific numeric data types with specific bit widths.
# Read the data with no imposed data types
df = pd.DataFrame(rows,
columns=[col.name for col in cur.description],
dtype=object)
# Assign specific int or float lengths to some fields
types = {'index': np.int32, 'id': np.int16, 'y': np.float32}
df = df.astype(types)
# Cast the Python datetime to a Pandas datetime
df['timestamp'] = pd.to_datetime(df.timestamp)
df.set_index('index').head(3)
| timestamp | id | name | x | y | |
|---|---|---|---|---|---|
| index | |||||
| 3456 | 2000-01-02 00:57:36 | 941 | Alice | -0.612 | -0.636485 |
| 3457 | 2000-01-02 00:57:37 | 1004 | Victor | 0.450 | -0.687718 |
| 3458 | 2000-01-02 00:57:38 | 980 | Quinn | 0.552 | 0.454158 |
We can verify those data types are used.
df.dtypes
index int32 timestamp datetime64[ns] id int16 name object x object y float32 dtype: object
Panda's "object" type hides the differences of the underlying classes of the Python objects stored. We can look at those more specifically.
pprint({repr(x): x.__class__.__name__
for x in df.reset_index().iloc[0]})
{"'Alice '": 'str',
'-0.636485': 'float32',
'0': 'int64',
'3456': 'int32',
'941': 'int16',
"Decimal('-0.612')": 'Decimal',
"Timestamp('2000-01-02 00:57:36')": 'Timestamp'}
For the most part, the steps for reading in SQL data in R are similar to those in Python. And so are the pitfalls around getting data types just right. We can see that the data types are the same rough approximations of the actual database types as Pandas produced. Obviously, in real code you should not specify passwords as literal values in the source code, but use some tool for secrets management.
%%capture --no-stdout err
%%R
require("RPostgreSQL")
drv <- dbDriver("PostgreSQL")
con <- dbConnect(drv, dbname = "dirty",
host = "localhost", port = 5432,
user = "cleaning", password = "data")
sql <- "SELECT id, name, x, y FROM dask_sample LIMIT 3"
data <- tibble(dbGetQuery(con, sql))
data
# A tibble: 3 x 4
id name x y
<int> <chr> <dbl> <dbl>
1 941 "Alice " -0.612 -0.636
2 1004 "Victor " 0.45 -0.688
3 980 "Quinn " 0.552 0.454
What is interesting to look at is that we might produce data frames that are not directly database tables (nor simply the first few rows, as in examples here), but rather some more complex manipulation or combination of that data. Joins are probably the most interesting case here since they take data from multiple tables. But grouping and aggregation is also frequently useful, and might reduce a million rows to a thousand summary descriptions, for example, which might be our goal.
%%capture --no-stdout err
%%R
sql <- "SELECT max(x) AS max_x, max(y) AS max_y,
name, count(name)
FROM dask_sample
WHERE id > 1050
GROUP BY name
ORDER BY count(name) DESC
LIMIT 12;"
# Here we simply retrieve a data.frame
# rather than convert to tibble
dbGetQuery(con, sql)
max_x max_y name count 1 0.733 0.7685581 Hannah 10 2 0.469 0.8493844 Norbert 10 3 0.961 0.7355076 Wendy 9 4 0.950 0.6730366 Quinn 8 5 0.892 0.8534938 Michael 7 6 0.772 0.9892333 Yvonne 7 7 0.958 0.8597920 Patricia 6 8 0.953 0.8659175 Ingrid 6 9 0.998 0.9807807 Oliver 6 10 0.050 0.5018596 Laura 6 11 0.399 0.8085724 Alice 5 12 0.604 0.8264008 Kevin 5
For the following example, I started with a data set that described Amtrak train stations in 2012. Many of the
fields
initially present were discarded, but some others were manipulated to illustrate some points. Think of this as
"fake
data" even though it is derived from a genuine data set. In particular, the column Visitors is
invented
whole cloth; I have never seen visitor count data, nor do I know if it is collected anywhere. It is just numbers
that
will have a pattern.
amtrak = pd.read_sql('bad_amtrak', engine)
amtrak.head()
| Code | StationName | City | State | Visitors | |
|---|---|---|---|---|---|
| 0 | ABB | Abbotsford-Colby | Colby | WI | 18631 |
| 1 | ABE | Aberdeen | Aberdeen | MD | 12286 |
| 2 | ABN | Absecon | Absecon | NJ | 5031 |
| 3 | ABQ | Albuquerque | Albuquerque | NM | 14285 |
| 4 | ACA | Antioch-Pittsburg | Antioch | CA | 16282 |
On the face of it—other than the telling name of the table we read in—nothing looks out of place. Let us look for problems. Notice that the tests below will, in a way, be anomaly detection, which is discussed in a later chapter. However, the anomalies we find are specific to SQL data typing.
String fields in RDBMS's are prone to truncation if specific character lengths are given. Modern database
systems
also have a VARCHAR or TEXT type for unlimited length strings, but often specific
lengths
are used in practice. To a certain degree, database operations can be more efficient with known text lengths, so
the
choice is not simple foolishness. But whatever the reason, you will find such fixed lengths frequently in
practice. In
particular, the StationName column is defined as CHAR(20). The question is: is that a
problem?
Knowing the character length will not automatically answer the question we care about. Perhaps Amtrak regulation requires a certain length of all station names. This is domain-specific knowledge that you may not have as a data scientist. In fact, the domain experts may not have it either, because it has not been analyzed or because rules have changed over time. Let us analyze the data itself.
Moreover, even if a database field is currently variable length or very long, it is quite possible that a column was altered over the life of a DB, or that a migration occurred. Unfortunately, multiple generations of old data that may each have been corrupted in their own ways can obscure detection.
One place you may encounter this problem data history is with dates in older data sets where two digit years
were
used. The "Y2K" issue had to be addressed two decades ago for active database systems—for example, I spent the
calendar year of 1998 predominantly concerned with this issue—but there remain some infrequently accessed legacy
data
stores that will fail on this ambiguity. If the character string '34' is stored in a column named
YEAR,
does it refer to something that happened in the 20th century or an anticipated future event a decade after this
book
is being written? Some domain knowledge is needed to answer this.
Some rather concise Pandas code can tell us something useful. A first step is cleaning the padding in the fixed length character field. The whitespace padding is not generally useful in our code. After that we can look at the length of each value, count the number of records per length, and sort by those lengths, to produce a histogram.
amtrak['StationName'] = amtrak.StationName.str.strip() hist = amtrak.StationName.str.len().value_counts().sort_index() hist
4 15
5 46
6 100
7 114
...
17 15
18 17
19 27
20 116
Name: StationName, Length: 17, dtype: int64
The pattern is even more striking if we visualize it. Clearly station names bump up against that 20 character width. This is not quite yet a smoking gun, but it is very suggestive.
hist.plot(kind='bar',
title="Lengths of Station Names")
plt.savefig('img/(Ch01)Lengths of Station Names')
We want to be careful not to attribute an underlying phenomenon as a data artifact. For example, in preparing this section, I started analyzing a collection of Twitter 2015 tweets. Those naturally form a similar pattern of "bumping up" against 140 characters—but I realized that they do this because of a limit in the accurate underlying data, not as a data artifact. However, the Twitter histogram curve looks similar to that for station names. I am aware that Twitter doubled its limit in 2018; I would expect an aggregated collection over time to show asymptotes at both 140 and 280, but as a "natural" phenomenon.
If the character width limit changed over the history of our data, we might see a pattern of multiple soft
limits.
These are likely to be harder to discern, especially if those limits are significantly larger than 20 characters
to
start with. Before we absolutely conclude that we have a data artifact rather than, for example, an
Amtrak
naming rule, let us look at the concrete data. This is not impractical when we start with a thousand rows, but
it
becomes more difficult with a million rows. Using Pandas' .sample() method is often a good way to
view a
random subset of rows matching some filter, but here we just display the first and last few.
amtrak[amtrak.StationName.str.len() == 20]
| Code | StationName | City | State | Visitors | |
|---|---|---|---|---|---|
| 28 | ARI | Astoria (Shell Stati | Astoria | OR | 27872 |
| 31 | ART | Astoria (Transit Cen | Astoria | OR | 22116 |
| 42 | BAL | Baltimore (Penn Stat | Baltimore | MD | 19953 |
| 50 | BCA | Baltimore (Camden St | Baltimore | MD | 32767 |
| ... | ... | ... | ... | ... | ... |
| 965 | YOC | Yosemite - Curry Vil | Yosemite National Park | CA | 28352 |
| 966 | YOF | Yosemite - Crane Fla | Yosemite National Park | CA | 32767 |
| 969 | YOV | Yosemite - Visitor C | Yosemite National Park | CA | 29119 |
| 970 | YOW | Yosemite - White Wol | Yosemite National Park | CA | 16718 |
116 rows × 5 columns
It is reasonable to conclude from our examined data that truncation is an authentic problem here. Many of the samples have words terminated in their middle at character length. Remediating it is another decision and more effort. Perhaps we can obtain the full texts as a followup; if we are lucky the prefixes will uniquely match full strings. Of course, quite likely, the real data is just lost. If we only care about uniqueness, this is likely not to be a big problem (the three letter codes are already unique). However, if our analysis concerns the missing data itself we may not be able to proceed at all. Perhaps we can decide in a problem-specific way that prefixes nonetheless are a representative sample of what we are analyzing.
A similar issue arises with numbers of fixed lengths. Floating-point numbers might lose desired precision, but
integers might wrap and/or clip. We can examine the Visitors column and determine that it stores a
16-bit
SMALLINT. Which is to say, it cannot represent values greater than 32,767. Perhaps that is more
visitors
than any single station will plausibly have. Or perhaps we will see data corruption.
max_ = amtrak.Visitors.max()
amtrak.Visitors.plot(
kind='hist', bins=20,
title=f"Visitors per station (max {max_})")
plt.savefig(f"img/(Ch01)Visitors per station (max {max_})")
In this case, the bumping against the limit is a strong signal. An extra hint here is the specific limit reached. It is one of those special numbers you should learn to recognize. Signed integers of bit-length N range from $-2^{N-1}$ up to $2^{N-1}-1$. Unsigned integers range from $0$ to $2^N$. 32,767 is $2^{16}-1$. However, for various programming reasons, numbers one (or a few) shy of a data type bound also frequently occur. In general, if you ever see a measurement that is exactly one of these bounds, you should take a second look and think about whether it might be an artifactual number rather than a genuine value. This is a good rule even outside the context of databases.
A possibly more difficult issue to address is when values wrap instead. Depending on the tools you use, large positive integers might wrap around to negative integers. Many RDBMS's—including PostgreSQL—will simply refuse transactions with unacceptable values rather than allow them to occur. But different systems vary. Wrapping on sign is obvious in the case of counts that are non-zero by their nature, but for values where both positive and negative numbers make sense, detection is harder.
For example, in this Pandas Series example, which is cast to a short integer type, we see values around positive and negative 15 thousand, both as genuine elements and as artifacts of a type cast.
ints = pd.Series(
[100, 200, 15_000, 50_000, -15_000, -50_000])
ints.astype(np.int16)
0 100 1 200 2 15000 3 -15536 4 -15000 5 15536 dtype: int16
In a case like this, we simply need to acquire enough domain expertise to know whether the out of bounds values that might wrap can ever be sensible measurements. I.e. is 50,000 reasonable for this hypothetical measure? If all reasonable observations are of numbers in the hundreds, wrapping at 32 thousand is not a large concern. It is conceivable that some reasonable value got there as a wrap from an unreasonable one; but wrong values can occur for a vast array of reasons, and this would not be an unduly large concern.
Note that integers and floating-point numbers only come, on widespread computer architectures, in sizes of 8, 16, 32, 64, and 128-bits. For integers those might be signed or unsigned, which would halve or double the maximum number representable. These maximum values representable within these different bit widths are starkly different from each other. A rule of thumb, if you can choose the integer representation, is to leave an order-of-magnitude padding from the largest magnitude you expect to occur. However, sometimes even an order-of-magnitude does not set a good bound on unexpected (but accurate) values.
For example, in our hypothetical visitor count, perhaps a maximum of around 20k was reasonably anticipated, but over the years, that got as high as 35k, leading to the effect we see in the plot (of hypothetical data). Allowing for 9,223,372,036,854,775,807 $(2^{63}-1)$ visitors to a station might have seemed like unecessary overhead to the initial database engineers. However, a 32-bit integer, with a maximum of 2,147,483,647 $(2^{31}-1)$ would have been a better choice, even though the actual maximum remains far larger than will ever be observed.
Let us turn now to some other data formats you are likely to work with, generally binary data formats, often used for scientific requirements.
Let a hundred flowers bloom; let a hundred schools of thought contend.
–Confucian saying
Concepts:
A variety of data formats that you may encounter can be used for holding tabular data. For the most part these do not introduce any special new cleanliness concerns that we have not addressed in earlier sections. Properties of the data themselves are discussed in later chapters. The data type options vary between storage formats, but the same kinds of general concerns that we discussed with RDBMS's apply to all of them. In the main, from the perspective of this book, these formats simply require somewhat different APIs to get at their underlying data, but all provide data types per column. The formats addressed herein are not an exhaustive list, and clearly new ones may arise or increase in significance after the time of this writing. But the principles of access should be similar for formats not discussed.
The closely related formats HDF5 and NetCDF (discussed below) are largely interoperable, and both provide ways of storing multiple arrays, each with metadata associated and also allowing highly dimensional data, not simply tabular 2-D arrays. Unlike with the data frame model, arrays within these scientific formats are of homogeneous type throughout. That is, there is no mechanism (by design) to store a text column and a numeric column within the same object, nor even numeric columns of different bit-widths. However, since they allow multiple arrays in the same file, full generality is available, just in a different way than within the SQL or data frame model.
SQLite (discussed below) is a file format that provides a relational database, consisting potentially of multiple tables, within a single file. It is extremely widely used, being present and used everywhere from on every iOS and Android device up to the largest supercomputer clusters. An interface for SQLite is part of the Python standard library and is available for nearly all other programming languages.
Apache Parquet (discussed below) is a column-oriented data store. What this amounts to is simply a way to store data frames or tables to disk, but in a manner that optimizes common operations that typically vectorize along columns rather than along rows. A similar philosophy motivates columnar RDBMS's like Apache Cassandra and MonetDB, both of which are SQL databases, simply with different query optimization possibilities. kdb+ is an older, and non-SQL, approach to a similar problem. PostgreSQL and MariaDBmariadb also both have optional storage formats that use column organization. Generally these internal optimizations are not direct concerns for data science, but Parquet requires its own non-SQL APIs.
mysql for
compatibility.
There are a number of binary data formats that are reasonably widely used, but I do not specifically discuss in this book. Many other formats have their own virtues, but I have attempted to limit the discussion to the handful that I feel you are most likely to encounter in regular work as a data scientist. Some additional formats are listed below, with characterization mostly adapted from their respective home pages. You can see in the descriptions which discussed formats they most resemble, and generally the identical data integrity and quality concerns apply as in the formats I discuss. Differences are primarily in performance characteristics: how big are the files on disk, how fast can they be read and written under different scenarios, and so on.
Apache Arrow is a development platform for in-memory analytics. It specifies a standardized language-independent columnar memory format for flat and hierarchical data, organized for efficient analytic operations on modern hardware.
bcolz provides columnar, chunked data containers that can be compressed either in-memory and on-disk. Column storage allows for efficiently querying tables, as well as for cheap column addition and removal. It is based on NumPy, and uses it as the standard data container to communicate with bcolz objects, but it also comes with support for import/export facilities to/from HDF5/PyTables tables and pandas dataframes.
Zarr provides classes and functions for working with N-dimensional arrays that behave like NumPy arrays but whose data is divided into chunks and each chunk is compressed. If you are already familiar with HDF5 then Zarr arrays provide similar functionality, but with some additional flexibility.
There is a slightly convoluted history of the Hierarchical Data Format, which was begun by the National Center for Supercomputing Applications (NCSA) in 1987. HDF4 was significantly over-engineered, and is far less widely used now. HDF5 simplified the file structure of HDF4. It consists of datasets, which are multidimensional arrays of a single data type, and groups, which are container structures holding datasets and other groups. Both groups and datasets may have attributes attached to them, which are any pieces of named metadata. What this, in effect, does is emulate a filesystem within a single file. The nodes or "files" within this virtual filesystem are array objects. Generally, a single HDF5 file will contain a variety of related data for working with the same underlying problem.
The Network Common Data Form (NetCDF) is a library of functions for storing and retrieving array data. The project itself is nearly as old as HDF, and is an open standard that was developed and supported by a variety of scientific agencies. As of its version 4, it supports using HDF5 as storage backend; earlier versions used some other file formats, and current NetCDF software requires continued support for those older formats. Occasionally NetCDF-4 files do enough special things with their contents that reading them with generic HDF5 libraries is awkward.
Generic HDF5 files typically have an extension of .h5, .hdf5, .hdf, or
.he5. These should all represent the same binary format, and other extensions occur sometimes too.
Some
corresponding extensions for HDF4 also exist. Oddly, even though NetCDF can consist of numerous underlying file
formats, they all seem standardized on the .nc extension.
Although I generally do not rely on GUI tools, in the case of viewing the fairly complex structure of HDF5, they can help. For example, a file from NASA's Earth Science Data collection is included in this book's sample data repository. Overall, NASA collects petabytes of Users can freely register to obtain data sets from NASA, which in aggregate is petabytes of information. This particular HDF5/NetCDF file contains datasets for surface pressure, vertical temperature profiles, surface and vertical wind profiles, tropopause pressure, boundary layer top pressure, and surface geopotential for a 98 minute period. In particular, some of the data is spatially 3-dimensional.
A view of a small portion of the data using the open source viewer HDF Compass illustrates some of the structure. The particular dataset viewed is one of 16 in the file. This DELP dataset is about pressure thickness, and contains both an array of 32-bit values and 8 attributes describing the dataset. You can see in the screenshot below that this particular GUI tool presents the 3rd dimension as a selection widget, and the first two dimensions in a tabular view.
Image: HDF Compass NASA Data
Within Python, there are two popular open source libraries for working with HDF5, PyTables and h5py. For working with NetCDF specifically, there is a library netcdf4-python as well. If you wish to read data from HDF5 files, and not to add NetCDF specific metadata, one of the general HDF5 tools is fine (h5py handles special metadata better than PyTables).
PyTables and h5py have moderately different attitudes. H5py stays close to HDF5 spec itself while PyTables attempts to provide a higher-level "Pythonic" interface. PyTables has the advantage that its data model borrows from, and acknowledges, a library for XML access that this author wrote way back in 2000; however, that advantage may be less relevant for general readers than for me personally. In the R world, the library rhdf5 is available.
In libraries for working with HDF5 data, a degree of laziness is allowed when dealing with large data sets. In the Python interfaces, data sets are virtualized NumPy arrays; importantly you can perform slice operations into these arrays, and only actually read into memory the indicated data. You may be dealing with terabytes of underlying information, but process or modify only megabytes at a time (or at all), with efficient reading and writing from regions of on disk arrays.
The names for data files used by NASA are verbose but contain detailed indication in the names themselves of the nature of the data sets within them. Let us open one file, and take a look at a summary of its data sets. We will show the data set name, its dimensions, its data type, its shape, and the 'units' attribute that these all happen to have. In general, attributes may have any names, but NASA has conventions about which to use.
import h5py
h5fname = ('data/earthdata/OMI-Aura_ANC-OMVFPITMET'
'_2020m0216t225854-o82929_v003'
'-2020m0217t090311.nc4')
data = h5py.File(h5fname, mode='r')
for name, arr in data.items():
print(f"{name:6s} | {str(arr.shape):14s} | "
f"{str(arr.dtype):7s} | {arr.attrs['units'][0]}")
DELP | (1494, 60, 47) | float32 | Pa PBLTOP | (1494, 60) | float32 | Pa PHIS | (1494, 60) | float32 | m+2 s-2 PS | (1494, 60) | float32 | Pa T | (1494, 60, 47) | float32 | K TROPPB | (1494, 60) | float32 | Pa U | (1494, 60, 47) | float32 | m s-1 U10M | (1494, 60) | float32 | m s-1 V | (1494, 60, 47) | float32 | m s-1 V10M | (1494, 60) | float32 | m s-1 lat | (1494, 60) | float32 | degrees_north lev | (47,) | int16 | 1 line | (1494,) | int16 | 1 lon | (1494, 60) | float32 | degrees_east sample | (60,) | int16 | 1 time | (1494,) | float64 | seconds since 1993-01-01 00:00:00
We can lazily create a memory view into only part of one of the dataset arrays. In the example, we have opened
in
read-only mode, but if we had opened using the 'r+' or 'a' modes we could change the
file.
Use the 'w' mode with extreme caution since it will overwrite an existing file. If the mode allows
modification on disk, calling data.flush() or data.close() will write any changes back
to
the HDF5 source.
Let us create a view of only a small section of the 3-dimensional V dataset. We are not particularly concerned here with understanding the domain of the data, but just demonstrating the APIs. In particular, notice that we have used a stride in one dimension to show that the general NumPy style of complex memory views is available. Only the data referenced is actually put into main memory while the rest stays on disk.
# A 3-D block from middle of DELP array middle = data['V'][::500, 10:12, :3] middle
array([[[17.032158 , 12.763597 , 3.7710803 ],
[16.53227 , 12.759642 , 4.1722884 ]],
[[ 4.003829 , -1.0843939 , -6.7918572 ],
[ 3.818467 , -1.0030019 , -6.6708655 ]],
[[-2.7798688 , 0.24923703, 20.513933 ],
[-2.690715 , 0.2226392 , 20.473366 ]]], dtype=float32)
If we modify the data in the view middle, it will be written back when we flush or close the
handle (if
not in read-only mode). We might also use our data slice for other computations or data science purposes. For
example,
perhaps such a selection acts as tensors that are input into a neural network. In a simpler case, perhaps we
simply
want to find some statistics or reduction/abstraction on the data.
middle.mean(axis=1)
array([[16.782215 , 12.76162 , 3.9716845 ],
[ 3.911148 , -1.0436978 , -6.7313614 ],
[-2.735292 , 0.23593812, 20.493649 ]], dtype=float32)
Working with HDF5 files in R—or most any other language—is generally similar to doing so from Python. Let us take a look with the R library rhdf5.
%%R -i h5fname library(rhdf5) h5ls(h5fname)
group name otype dclass dim 0 / DELP H5I_DATASET FLOAT 47 x 60 x 1494 1 / PBLTOP H5I_DATASET FLOAT 60 x 1494 2 / PHIS H5I_DATASET FLOAT 60 x 1494 3 / PS H5I_DATASET FLOAT 60 x 1494 4 / T H5I_DATASET FLOAT 47 x 60 x 1494 5 / TROPPB H5I_DATASET FLOAT 60 x 1494 6 / U H5I_DATASET FLOAT 47 x 60 x 1494 7 / U10M H5I_DATASET FLOAT 60 x 1494 8 / V H5I_DATASET FLOAT 47 x 60 x 1494 9 / V10M H5I_DATASET FLOAT 60 x 1494 10 / lat H5I_DATASET FLOAT 60 x 1494 11 / lev H5I_DATASET INTEGER 47 12 / line H5I_DATASET INTEGER 1494 13 / lon H5I_DATASET FLOAT 60 x 1494 14 / sample H5I_DATASET INTEGER 60 15 / time H5I_DATASET FLOAT 1494
You may notice that the order of dimensions is transposed in R versus Python, so we have to account for that in
our
selection of a region of interest. However, generally the operation of slicing in R is very similar to that in
NumPy.
The function h5save() is used to write data that was modified back to disk.
%%R -i h5fname V = h5read(h5fname, 'V') V[1:2, 10:12, 10:11]
, , 1 [,1] [,2] [,3] [1,] 17.69524 17.23481 16.57238 [2,] 12.46370 12.44905 12.47155 , , 2 [,1] [,2] [,3] [1,] 17.71876 17.25898 16.56942 [2,] 12.42049 12.40599 12.43139
The NASA data shown does not use group hierarchies, only top-level datasets. Let us look at a toy data collection that nests groups and datasets.
make_h5_hierarchy() # initialize the HDF5 file
f = h5py.File('data/hierarchy.h5', 'r+')
dset = f['/deeply/nested/group/my_data']
print(dset.shape, dset.dtype)
(10, 10, 10, 10) int32
We see that we have a 4-dimensional array of integer data. Perhaps some metadata description was attached to it
as
well. Let us also view—and then modify—some section of the data since we have opened in 'r+' mode.
After
we change the data, we can write it back to disk. We could similarly change or add attributes in a regular
dictionary
style; for instance:
dset.attrs[mykey] = myvalue
Let us show a slice from the dataset.
for key, val in dset.attrs.items():
print(key, "→", val)
print()
print("Data block:\n", dset[5, 3, 2:4, 8:])
author → David Mertz citation → Cleaning Data Book shape_type → 4-D integer array Data block: [[-93 -53] [ 18 -37]]
Now we modify the same slice of data we displayed, then close the file handle to write it back to disk.
dset[5, 3, 2:4, 8:] = np.random.randint(-99, 99, (2, 2)) print(dset[5, 3, 2:4, 8:]) f.close() # write change to disk
[[-45 -76] [-96 -21]]
We can walk the hierarchy in Python's h5py package, but it is somewhat manual to loop through
paths. R's
rhdf5 provides a nice utility function h5ls() that lets us see more of the structure
of this
test file.
%%R
library(rhdf5)
h5ls('data/hierarchy.h5')
group name otype dclass dim 0 / deeply H5I_GROUP 1 /deeply nested H5I_GROUP 2 /deeply/nested group H5I_GROUP 3 /deeply/nested/group my_data H5I_DATASET INTEGER 10 x 10 x 10 x 10 4 /deeply path H5I_GROUP 5 /deeply/path elsewhere H5I_GROUP 6 /deeply/path/elsewhere other H5I_DATASET INTEGER 20 7 /deeply/path that_data H5I_DATASET FLOAT 5 x 5
In essence, SQLite is simply another RDBMS from the point of view of a data scientist. For a developer or
systems
engineer, it has some special properties, but for readers of this book, you will get data from an SQLite file
via SQL
queries. Somewhat similarly to HDF5, an SQLite file—often given extensions .sqlite,
.db, or
.db3 (but not as standardized as with some file types)—can contain many tables. In SQL, we
automatically
get joins and subqueries to combine data from multiple tables, whereas there is no similar standard about
combining
data from multiple HDF5 datasets.
The SQLite3 data format and server is extremely efficient, and queries are usually fast. As with other SQL databases, it operates with atomic transactions that succeed or fail in their entirety. This prevents a database from reaching a logically inconsistent state. However, it does not have a concurrent access model. Or rather, it does not allow multiple simultaneous writers to a common database in the way that server-based RDBMSs do. Many reader clients may open the same file simultaneously without difficulty; it only bogs down when many clients wish to perform write transactions. There are ways to address this situation, but they are outside the scope of this particular book.
An important advantage of SQLite over other RDBMS's is that distributing the single file that makes up the database is dead simple. With other systems, you need to add credentials, and firewall rules, and the like, to give new users access; or alternately you need to export the needed data to another format, typically CSV, that is both slow and somewhat lossy (i.e. data types).
Data typing in SQLite is something of a chimera. There are few basic data types, which we will discuss. However, unlike nearly every other SQL database, SQLite carries data types per value, not per column. This would seem to run into the same fragility that was discussed around spreadsheets, but in practice it is far less of a problem than with those. One reason the types-per-value is not as much of a concern is because of the interface used to populate them; it is highly unusual to edit individual values in SQLite interactively, and far more common to issue programmatic SQL commands to INSERT or UPDATE many rows with data from a common source.
However, apart from the data types, SQLite has a concept called type affinity. Each column is given a
preferred type that does not prevent other data types from occurring, but does nudge the preference
toward
the affinity of the column. We can run the tool sqlite from the command line to get to the
interactive
SQLite prompt. For example (adapted from SQLite documentation):
sqlite> CREATE TABLE mytable(a SMALLINT, b VARCHAR(10), c REAL);
sqlite> INSERT INTO mytable(a, b, c) VALUES('123', 456, 789);
Here a row will be inserted with an integer in the a column, TEXT in the b column,
and a
floating-point in the c column. SQL syntax itself is loosely typed, but the underlying database
makes
type/casting decisions. This is true of other RDBMS's too, but those are stricter about column data types. So we
can
also run this in SQLite, which will fail with other databases:
sqlite> INSERT INTO mytable(a, b, c) VALUES('xyz', 3.14, '2.71');
Let us see what results:
sqlite> SELECT * FROM mytable; 123|456|789.0 xyz|3.14|2.71
The SQLite interactive shell does not make data types entirely obvious, but running a query in Python will do so.
import sqlite3
db = sqlite3.connect('data/affinity.sqlite')
cur = db.cursor()
cur.execute("SELECT a, b, c FROM t1")
for row in cur:
print([f"{x.__class__.__name__} {x}" for x in row])
['int 123', 'str 456', 'float 789.0'] ['str xyz', 'str 3.14', 'float 2.71']
Column a prefers to hold an integer if it is set with something it can interpret as an integer,
but will
fall back to a more general data type if required. Likewise, column c prefers a float, and it can
interpret either an unquoted integer or a float-like string as such.
The actual data types in SQLite are exclusively NULL, INTEGER, REAL, TEXT, and BLOB. However, most of
the
type names in other SQL databases are aliases for these simple types. We see that in the example, where
VARCHAR(10) is an alias for TEXT and SMALLINT is an alias for
INTEGER. REAL values are always represented as 64-bit floating-point numbers. Within INTEGER
values, bit
lengths of 1, 2, 3, 4, 6, or 8 are chosen for storage efficiency. There is no datetime type in SQLite storage,
but
time-oriented SQL functions are happy to accept any of TEXT (ISO-8601 strings), REAL (days since November 24,
4714
B.C), or INTEGER (seconds since 1970-01-01T00:00:00).
The overall takeaway for working with SQLite databases is that possibly a little extra care is needed in double checking your data types when reading data, but for the most part you can pretend it is strongly typed per column. Truncation, clipping, and wrap-around issues will not occur. There is no actual decimal data type, but only aliases; for data science—versus accounting or finance—this is rarely a concern. But usual caveats about floating-point rounding issues will apply.
The Parquet format grew out of the Hadoop ecosystem, but at heart is simply an optimized, column-oriented file format for storing table-like data. Parquet has a type system that focuses on numeric types. It is not quite as simplified as SQLite, but also eschews providing every possible bit length, as NumPy or C/C++ do, for example. All integer types are signed. Everything that is not numeric is a byte-array that is cast for the needed purpose at the application level (i.e. not the storage format level).
Having grown out of Hadoop tools, Parquet is especially well optimized for parallel computation. A Parquet
"file" is
actually a directory containing a number of data files, with a _metadata file in that directory
describing the layout and other details.
%%bash ls -x data/multicsv.parq
_common_metadata _metadata part.0.parquet part.1.parquet part.10.parquet part.11.parquet part.12.parquet part.13.parquet part.14.parquet part.15.parquet part.16.parquet part.17.parquet part.18.parquet part.19.parquet part.2.parquet part.20.parquet part.21.parquet part.22.parquet part.23.parquet part.24.parquet part.25.parquet part.26.parquet part.27.parquet part.28.parquet part.29.parquet part.3.parquet part.4.parquet part.5.parquet part.6.parquet part.7.parquet part.8.parquet part.9.parquet
Sometimes the file system is a parallel and distributed system such as Hadoop File System (HDS) that further supports computational efficiency on large data sets. In such case, Parquet does various clever sharding of data, efficient compression (using varying strategies), optimization of contiguous reads, and has been analyzed and revised to improve its typical use cases, for both speed and storage size.
Some of the tools or libraries supporting Parquet are Apache Hive, Cloudera Impala, Apache Pig, and Apache Spark, all of which live in the parallel computation space. However, there are available interfaces for Python and R as well (and other languages). Many of the higher level tools address Parquet data with an SQL layer.
For Python, the libraries pyarrow and fastparquet provide a direct interface to the file format. While these libraries are general, they are designed primarily to translate Parquet data into data frames (usually Pandas, sometimes Dask, Vaex, or others). Within the R world, sparklyr is an interface into Spark, but requires a running Spark instance (a local installation is fine). The arrow package is a direct reader, similar to the Python libraries.
In general, if you are working with genuinely big data, the Hadoop or Spark tools—accompanied by appropriate computing clusters—are a good choice. Dask is an approach to parallelism on Python, which is very good; other approaches like MPI are available for R, Python, and many other languages. However, Hadoop and Spark are the tools to which the most attention has been paid in regard to efficient and large scale parallel computation.
Even if you only need to worry about medium sized data (hundred of thousands to millions of rows) rather than big data (hundreds of millions to billions of rows), Parquet is still a fast format to work with. Moreover, it has the generally desirable property of typing data by column that makes data at least one small step closer to being clean and tidy.
As an example, let us read the medium sized data set we generated earlier with Dask. Both Pandas and Dask will use either pyarrow or fastparquet, depending on what is installed.
pd.read_parquet('data/multicsv.parq/')
| timestamp | id | name | x | y | |
|---|---|---|---|---|---|
| index | |||||
| 0 | 2000-01-01 00:00:00 | 979 | Zelda | 0.802163 | 0.166619 |
| 1 | 2000-01-01 00:00:01 | 1019 | Ingrid | -0.349999 | 0.704687 |
| 2 | 2000-01-01 00:00:02 | 1007 | Hannah | -0.169853 | -0.050842 |
| 3 | 2000-01-01 00:00:03 | 1034 | Ursula | 0.868090 | -0.190783 |
| ... | ... | ... | ... | ... | ... |
| 86396 | 2000-01-10 23:59:56 | 998 | Jerry | 0.589575 | 0.412477 |
| 86397 | 2000-01-10 23:59:57 | 1011 | Yvonne | 0.047785 | -0.202337 |
| 86398 | 2000-01-10 23:59:58 | 1053 | Oliver | 0.690303 | -0.639954 |
| 86399 | 2000-01-10 23:59:59 | 1009 | Ursula | 0.228775 | 0.750066 |
2592000 rows × 5 columns
We could distribute the below read using dask.dataframe and just the same syntax, i.e.
dd.read_parquet(...). For large data sets the this could keep the inactive segments out of core and
distribute work over all the cores on the local machine. However, for medium to small data like this, Pandas is
faster
in avoiding the coordination overhead.
Although we have utilized the concept of data frames already, using both Python with Pandas and R with tibbles, it is worth looking at just what the underlying abstraction consists of. We will briefly look at a number of different data frame implementations in varying programming languages to understand what they have in common (with is a lot).
Whenever you set out to do something, something else must be done first.
—Murphy's (First) Corollary
Concepts:
A large number of libraries across almost as many programming languages support the data frame abstraction. Most data scientists find this abstraction to be powerful, and even their preferred way of processing data. Data frames allow an easy expression of many of the same fundamental concepts or operations as does SQL, but within the particular programming language and memory space of the rest of their program. SQL—even when it actually addresses a purely local database such as SQLite—is always more of a "remote fetch" than interactive exploration that data frames allow.
These operations consist, in the main, of filtering, grouping, aggregation, sorting, and vectorized function application. Generally, all data frame libraries allow for a "fluent" programming style that chains together these operations in some order to produce a final result; that final (or at least working) result is usually itself either a data frame or a scalar value. Sometimes a visualization is relevant for such a processed result, and most data frame tools integrate seamlessly with visualization libraries.
The goal, of course, of these fluent chained operations is to describe a reproducible work flow. Exploration of various data modifications can be built up step by step, with intermediate results often providing hints that you might have gone wrong or a degree of reassurance that your path is correct. At the end of that exploration, you will have an expression of a composite transformation of data that can be reused with new data from the domain and problem you are addressing. Comments in code and accompanying these chains, or pipelines, always make life easier for both you and other readers of code.
Those libraries that are distributed and/or out-of-core allow working with large data sets rather seamlessly. Which is to say that the data frame abstraction scales almost unboundedly, even if particular libraries have some rough limits. In this section, I will present similar code using a number of data frame libraries, commenting briefly on the strengths, weaknesses, and differences among them. This book generally utilizes Python with Pandas, and to a somewhat lesser extent R with tibbles. We will see the conceptual and usage similarity of those libraries with other libraries in Python/R (Vaex, data.table), and even with other programming languages such as Scala/Spark or bash with coreutils. Many data scientists use Spark, in particular; whatever specific tools you use, the concepts throughout should translate easily, especially where data frame approaches are available.
Most of the code in this book will use Pandas. Python is, as of this writing, the most widely used language for data science, and Pandas is, by a large margin, its most widely used data frame library. In fact, several of the "competing" libraries themselves utilize Pandas as an internal component. However, in this section, I would like to illustrate and emphasize how similar all of these libraries are. For that purpose, I am going to perform the same task using a number of these libraries.
There are a great many operations and pipelines, many quite complex, that can be accomplished with data frames. This brief section is not a tutorial on any of the specific libraries, but only a glimpse into the shared style of expressing data manipulation and the smaller differences among the different tools.
With each data frame library, we will do the following:
x and yynamexMean_xAs a starting point, I would like to illustrate a pipeline of steps using the distributed computing framework, Spark, and its native programming language, Scala. Bindings into Spark from Python, R, and other languages also exist, but incur a certain degree of translation overhead that slow operations. This pipeline takes the sample Dask data set shown in other examples in this chapter, and performs all of the basic operations mentioned on the data set.setup
The next few lines were run inside the Spark shell. For composition of this book, a local instance of Hadoop and Spark were running, but this could as easily be a connection to a remote cluster. Upon launch you will see something similar to this:
Spark context Web UI available at http://popkdm:4040
Spark context available as 'sc' (master = local[*], app id = local-1582775303458).
Spark session available as 'spark'.
Welcome to
____ __
/ __/__ ___ _____/ /__
_\ \/ _ \/ _ `/ __/ '_/
/___/ .__/\_,_/_/ /_/\_\ version 2.4.5
/_/
Using Scala version 2.11.12 (OpenJDK 64-Bit Server VM, Java 11.0.6)
Type in expressions to have them evaluated.
Type :help for more information.In the shell, we can read in the collection of CSV files in a common directory. Many other data sources are
likewise
available under a similar interface. We allow inference of data types and use of the column headers to name
fields.
The pipe symbols (|) are simply part of the Spark shell interface to indicate a continuation line;
they
are not themselves the Scala code.
scala> val df = spark.read. // Local file or Hadoop resource
| options(Map("inferSchema"->"true","header"->"true")).
| csv("data/multicsv/") // Directory of multiple CSVs
df: org.apache.spark.sql.DataFrame = [
timestamp: timestamp, id: int ... 3 more fields]
The fluent code below simply performs the intended steps in order.
scala> df. // Working with loaded DataFrame
| filter($"x" > ($"y" + 1)). // x more than y+1 (per row)
| groupBy($"name"). // group together same name
| agg(avg($"x") as "Mean_x"). // mean within each group
| sort($"Mean_x"). // order data by new column
| show(5)
+------+------------------+
| name| Mean_x|
+------+------------------+
| Ray|0.6625697073245446|
|Ursula|0.6628107271270461|
|Xavier|0.6641165295855926|
| Wendy|0.6642381725604264|
| Kevin| 0.664836301676443|
+------+------------------+
only showing top 5 rowsA number of libraries either emulate the Pandas API or directly utilize it as a dependency. Dask and Modin both directly wrap Pandas, and partition one native DataFrame into many separate Pandas DataFrames. A method on the native DataFrame is usually dispatched to the underlying corresponding Pandas method per-DataFrame. Although Modin can use either Dask or Ray as its parallel/cluster execution back-end, Modin differs from Dask in being eager in its execution model.
Dask is a general purpose execution back-end, with its dask.dataframe subpackage being only one
component. Much of what Dask does is similar to the library Ray, which Modin may also use if desired (benchmarks
as of
this writing mildly favor Ray, depending on use-case). Most Pandas API method calls in Dask initially only build
a
directed-acyclic graph (DAG) of the required operations. Computation is only performed when the
.compute() method of a built DAG is called. The example below uses Dask, but it would look exactly
the
same with Modin except for the final .compute() and an initial
import modin.pandas as pd.
cuDF is another library that follows Pandas' API very closely, but it executes methods on CUDA GPUs. Since the underlying execution is on an entirely different kind of chip architecture, cuDF does not share code with Pandas, nor wrap Pandas. But almost all API calls will be identical, but often vastly faster if you have a recent CUDA GPU on your system. Like Pandas and Modin, cuDF is eager in its execution model.
import dask.dataframe as dd
dfd = dd.read_csv('data/multicsv/*.csv', parse_dates=['timestamp'])
The operations in the Pandas style below look very similar to those in Spark. The accessor .loc
overloads several selection styles, but a predicate filter is such a permitted one. Another is used on the same
line
to select columns, i.e. a sequence of names. Grouping is nearly identical, other than the capitalization of the
method
name. Pandas even has an .agg() method to which we could pass a mean function or the string
'mean'; we just chose the shortcut. Columns are not automatically renamed in aggregation, so we do
that
to match more precisely. Instead of sorting and showing, we take the 5 smallest in a single method. In effect,
the
conceptual elements are identical, and spelling varies only mildly.
(dfd
.loc[dfd.x > dfd.y+1, # Row predicate
['name', 'x']] # Column list
.groupby("name") # Grouping column(s)
.mean() # Aggregation
.rename(columns={'x': 'Mean_x'}) # Naming
.nsmallest(5, 'Mean_x') # Selection by order
).compute() # Concretize
| Mean_x | |
|---|---|
| name | |
| Ray | 0.662570 |
| Ursula | 0.662811 |
| Xavier | 0.664117 |
| Wendy | 0.664238 |
| Kevin | 0.664836 |
Vaex is a Python library completely independent of Pandas, but that uses a largely similar API. A fair amount of code will just work identically with either style of data frame, but enough will not that you cannot simply drop in one for the other. The philosophy of Vaex is somewhat different from Pandas. On the one hand, Vaex emphasizes lazy computation and implicit parallelism; expressions are eagerly evaluated, but with attention to not touching those portions of data that are not needed for a given operation. This goes hand-in-hand with the mostly out-of-core operation. Rather than read data into memory, Vaex memory maps the data on disk, only loading those parts required for an operation.
Vaex consistently avoids making data copies, in effect expressing selections as views. It has a concept of expressions and of virtual columns. For example, a computation on several columns, even if assigned to a new column, does not use any significant new memory since only the functional form is stored rather than the data. Only when that data is needed is the computation performed, and only for those rows affected. The overall result is that Vaex can be very fast on large data sets; however, Vaex parallelizes only over multiple cores on one machine, not over clusters of machines.
Because of its memory-mapped approach, Vaex does not really want to deal directly with CSV files internally.
Unlike
serialized Feather or HDF5, which put each datum at a predictable location on disk, CSV is inherently ragged in
layout
on disk. While a .read_csv() method will read a single file into memory, for working with a family
of
CSVs in a directory, you will want to convert them to a corresponding family of HDF5 files. Fortunately, the
method
.read_csv_and_convert() does this automatically for you. The result is that the first time you read
such
a collection, the conversion takes a while, but subsequent opens utilize the existing HDF5 files and open
instantly
(no actual read into memory, just memory maps).
import vaex
dfv = vaex.read_csv_and_convert('data/multicsv/*.csv', copy_index=False)
vaex.__version__
{'vaex': '3.0.0',
'vaex-core': '2.0.3',
'vaex-viz': '0.4.0',
'vaex-hdf5': '0.6.0',
'vaex-server': '0.3.1',
'vaex-astro': '0.7.0',
'vaex-jupyter': '0.5.2',
'vaex-ml': '0.9.0',
'vaex-arrow': '0.5.1'}
Another difference from Pandas is that Vaex data frames are tidy (as described at the start of this chapter). Many operations on Pandas rely on their row index, which might even be a hierarchical index comprising multiple nested columns. The "index," such as it is, in Vaex is simply the row number. You can do filtering, and grouping, and sorting, and so on, but always based on regular columns. This philosophy is shared with tibble and data.table in R, both of which reject that aspect of the older data.frame.
print(
dfv
[dfv.x > dfv.y + 1] # Predicate selection of rows
[['name', 'x']] # List selection of columns
.groupby('name') # Grouping
.agg({'x': 'mean'}) # Aggregation
.sort('x') # Sort (Vaex does not have .nsmallest() method)
.head(5) # First 5
)
# name x 0 Ray 0.66257 1 Ursula 0.662811 2 Xavier 0.664117 3 Wendy 0.664238 4 Kevin 0.664836
Let us remove those temporary HDF5 files for discussion of libraries other than Vaex.
%%bash rm -f data/multicsv/*.hdf5
Let us now turn to analogous data frame options within R.
In the Tidyverse, tibbles are the preferred data frame objects, and dplyr is an associated
library
for—often chained—pipelined data manipulations. The way that dplyr achieves a fluent style is not based on
chained
method calls. Indeed, object-oriented programming is rarely used in R in general. Instead, dplyr relies on the
"pipe"
operator (%>%) which treats the result of the prior expression as the first argument to the next
function called. This allows for rewriting compact but deeply nested expressions, such as the following:
round(exp(diff(log(x))), 1)
In fluent style this becomes:
x %>% log() %>% diff() %>% exp() %>% round(1)
First we can read in the collection of CSV files that was generated earlier. The 2.5 million total rows in this data are still medium sized data, but the patterns in the below code could be applied to big data.
%%R
files <- dir(path = "data/multicsv/", pattern = "*.csv", full.names = TRUE)
read_csv_quiet <- function(file) {
read_csv(file, col_types = cols("T", "n", "f", "n", "n"), progress = FALSE) }
data <- files %>%
# read_csv() on each file, reduce to one DF with rbind
map(read_csv_quiet) %>%
# If this were genuinely large data, we would process each file individually
reduce(rbind)
data
# A tibble: 2,592,000 x 5 timestamp id name x y <dttm> <dbl> <fct> <dbl> <dbl> 1 2000-01-01 00:00:00 979 Zelda 0.802 0.167 2 2000-01-01 00:00:01 1019 Ingrid -0.350 0.705 3 2000-01-01 00:00:02 1007 Hannah -0.170 -0.0508 4 2000-01-01 00:00:03 1034 Ursula 0.868 -0.191 5 2000-01-01 00:00:04 1024 Ingrid 0.0838 0.109 6 2000-01-01 00:00:05 955 Ingrid -0.757 0.308 7 2000-01-01 00:00:06 968 Laura 0.230 -0.913 8 2000-01-01 00:00:07 945 Ursula 0.265 -0.271 9 2000-01-01 00:00:08 1020 Victor 0.512 -0.481 10 2000-01-01 00:00:09 992 Wendy 0.862 -0.599 # … with 2,591,990 more rows
The dplyr pipes into functions that filter, modify, group, and aggregate data look nearly identical to the chained methods used in other data frame libraries. A few function names are slightly different than in other libraries, but the steps performed are identical.
%%R
info <- capture.output(
# --- hide nonsense on STDERR
summary <- data %>%
filter(x > y+1) %>% # Predicate selection of rows
select(name, x) %>% # Selection of columns
group_by(name) %>% # Grouping
# Aggregation and naming
summarize(Mean_x = mean(x)) %>%
arrange(Mean_x) %>% # Sort data
head(5) # First 5
# --- end of block for capture.output
,type = "message")
summary
`summarise()` ungrouping output (override with `.groups` argument) # A tibble: 5 x 2 name Mean_x <fct> <dbl> 1 Ray 0.663 2 Ursula 0.663 3 Xavier 0.664 4 Wendy 0.664 5 Kevin 0.665
Outside the Tidyverse, the main approach to working with tabular data in modern R is data.table. This is a replacement for the older, but standard, R data.frame. I do not separately discuss data.frame in this book since new code should always prefer either tibbles or data.tables.
Unlike most other approaches to data frames, data.table does not use a fluent or chained style. Instead, it
uses an
extremely compact general form of DT[i, j, by] that captures a great many of the
manipulations
possible. Not every collection of operation can be expressed in a single general form, but a great many of them
can.
Moreover, because data.table is able to optimize over the entire general form, it can often be significantly
faster on
large data sets than those libraries performing operations in a sequenced manner.
Each element of the general form may be omitted to mean "everything." If used, the i is an
expression
describing the rows of interest; often this i will consist of several clauses joined by logic
connectors
& (and), | (or), and ! (not). Row ordering may also be imposed within
this
expression (but not on derived columns). For example:
dt[(id > 999 | date > '2020-03-01') & !(name == "Lee")]
The column selector j can refer to columns, including derived columns.
dt[ , .(id, pay_level = round(log(salary), 1)]
Finally, the by form is a grouping description that allows for calculations per row subset. Groups
can
follow either categorical values or computed cuts.
dt[, mean(salary), cut(age, quantile(age, seq(0,100,10)))]
Putting those forms together, we can produce the same summary as with other data frame libraries. However, the final ordering has to be performed as a second step.
%%R
info <- capture.output(
# --- hide nonsense on STDERR
library(data.table)
# --- end of block for capture.output
,type = "message")
dt <- data.table(data)
summary <- dt[
i = x > y + 1, # Predicate selection of rows
# Aggregation and naming
j = .(Mean_x = mean(x)),
by = .(name)] # Grouping
# Sort data and first 5
summary[order(Mean_x), .SD[1:5]]
name Mean_x 1: Ray 0.6625697 2: Ursula 0.6628107 3: Xavier 0.6641165 4: Wendy 0.6642382 5: Kevin 0.6648363
For readers who are accustomed to performing pipelined filtering and aggregation at the command line, the piped or fluent style used by data frames will seem very familiar. In fact, it is not difficult to replicate our example using command-line tools. The heavy lifter here is awk, but the code it uses is very simple. Conceptually, these steps exactly match those we used in data frame libraries. The small tools that combine, using pipes, under the Unix philosophy, can naturally replicate the same basic operations used in data frames.
%%bash
COND='{if ($4 > $5+1) print}'
SHOW='{for(j in count) print j,sum[j]/count[j]}'
AGG='{ count[$1]++; sum[$1]+=$2 }'" END $SHOW"
cat data/multicsv/*.csv | # Create the "data frame"
grep -v ^timestamp | # Remove the headers
awk -F, "$COND" | # Predicate selection
cut -d, -f3,4 | # Select columns
awk -F, "$AGG" | # Aggregate by group
sort -k2 | # Sort data
head -5 # First 5
Ray 0.66257 Ursula 0.662811 Xavier 0.664117 Wendy 0.664238 Kevin 0.664836
Jeroen Janssens wrote a delightful book entitled [Data Science at the Command Line](https://www.datascienceatthecommandline.com/) that is both wonderfully written and freely available online. You should also buy the printed or ebook edition to support his work. In this subsection, and at various places in this book, I make only small gestures in the direction of the types of techniques that book talks about in detail.
The data frame and fluent programming style is a powerful idiom, and is especially widely used in data science. Every one of the specific libraries I discuss are excellent choices with equivalent power. Which fits you best is largely a matter of taste, and perhaps of what your colleagues use.
Putting together much of what we have learned in this chapter, the below exercises should allow you to utilize the techniques and idioms you have read about.
An Excel spreadsheet with some brief information on awards given to movies is available at:
In a more fleshed out case, we might have data for many more years, more types of awards, more associations that grant awards, and so on. While the organization of this spreadsheet is much like a great many you will encounter "in the wild," it is very little like the tidy data we would rather work with. In the simple example, only 63 data values occur, and you could probably enter them into the desired structure by hand as quickly as coding the transformations. However, the point of this exercise is to write programming code that could generalize to larger data sets of similar structure.
Image: Film Awards Spreadsheet
Your task in this exercise is to read this data into a single well-normalized data frame, using whichever language and library you are most comfortable with. Along the way, you will need to remediate whatever data integrity problems you detect. As examples of issues to look out for:
In thinking about a good data frame organization, think of what the independent and dependent variables are. In each year, each association awards for each category. These are independent dimensions. A person name and a film name are slightly tricky since they are not exactly independent, but at the same time some awards are to a film and others to a person. Moreover, one actor might appear in multiple films in a year (not in this sample data; but do not rule it out). Likewise, at times multiple films have used the same name at times in film history. Some persons are both director and actor (in either the same or different films).
Once you have a useful data frame, use it to answer these questions in summary reports:
An SQLite database with roughly the same brief information as in the prior spreadsheet is available at:
However, the information in the database version is relatively well normalized and typed. Also, additional information has been included on a variety of entities included in the spreadsheet. Only slightly more information is included in this schema than in the spreadsheet, but it should be able to accommodate a large amount of data on films, actors, directors, and awards, and the relationships among those data.
sqlite> .tables actor award director org_name
As was mentioned in the prior exercise, the same name for a film can be used more than once, even by the same director. For example Abel Gance, used the title J'accuse! for both his 1919 and 1938 films with connected subject matter.
sqlite> SELECT * FROM director WHERE year < 1950;
Abel Gance|J'accuse!|1919
Abel Gance|J'accuse!|1938Let us look at a selection from the actor table, for example. In this table we have a column
gender to differentiate beyond name. As of this writing, no transgender actor has been nominated
for a
major award both before and after a change in gender identity, but this schema allows for that possibility. In
any
case, we can use this field to differentiate the "actor" versus "actress" awards that many organizations grant.
sqlite> .schema actor CREATE TABLE actor (name TEXT, film TEXT, year INTEGER, gender CHAR(1)); sqlite> SELECT * FROM actor WHERE name="Joaquin Phoenix"; Joaquin Phoenix|Joker|2019|M Joaquin Phoenix|Walk the Line|2006|M Joaquin Phoenix|Hotel Rwanda|2004|M Joaquin Phoenix|Her|2013|M Joaquin Phoenix|The Master|2013|M
The goal in this exercise is to create the same tidy data frame that you created in the prior exercise, and answer the same questions that were asked there. If some questions can be answered directly with SQL, feel free to take that approach instead. For this exercise, only consider awards for the years 2017, 2018, and 2019. Some others are included in an incomplete way, but your reports are for those years.
sqlite> SELECT * FROM award WHERE winner="Frances McDormand"; Oscar|Best Actress|2017|Frances McDormand GG|Actress/Drama|2017|Frances McDormand Oscar|Best Actress|1997|Frances McDormand
All models are wrong, but some models are useful.
–George Box
Topics: Delimited Files; Spreadsheet Dangers; RDBMS; HDF5; Data Frames.
This chapter introduced the data formats that make up the large majority of all the structured data in the world. While I do not have hard data, exactly, on this breakdown of data volume—nor can anyone, apart perhaps, from some three letter agencies specializing in bulk data acquisition—I still feel like it is a safe assertion. Between all the scientific data stored in HDF5 and related formats, all the business data stored in spreadsheets, all the transactional data stored in SQL databases, and everything exported from almost everywhere to CSV, this makes up almost everything a working data scientist encounters on a regular basis.
In presenting formats, we addressed the currently leading tools for ingestion of those data sources in several languages. The focus throughout this book will remain on Python and R, which are the main programming languages for data science. Perhaps that will change in the future, and almost certainly some new libraries will arise for addressing this huge bulk of data in faster and more convenient ways. Even so, most of the conceptual issues about the strengths and limits of formats—concerns largely about data types and storage artifacts—will remain for those new languages and libraries. Only spelling will change mildly.
An extended, but nonetheless dramatically incomplete, discussion looked at the data frame abstraction used in a great many tools. Here again, new variations may arise, but I am confident that the general abstraction will be the primary one used in data science for several decades after this writing. In presenting a number of slightly different libraries, I have only scratched the surface of any one of them. In fact, even if this entire chapter was about just one of the mentioned libraries, it would be incomplete compared with those excellent books that spend their whole length discussing one particular data frame library. Nonetheless, I hope that this introduction to thinking about data processing problems in terms of the steps of filtering, grouping, aggregation, naming, and ordering will serve readers well in their articulation of many ingestion tasks.
One limit of the data frame abstraction that we used in reading all the formats discussed in this chapter is that none look at data streaming in any meaningful way. For the most part, data science needs are not streaming needs, but occasionally they overlap. If your needs lie at that particular edge, check the documentation for streaming protocols like ActiveMQ, RabbitMQ, and Kafka (among others); but your concern will not chiefly be in the data formats themselves, but rather in event processing, and in evolving detection of anomalies and bad data, such as is discussed in later chapters 4 and 5, and perhaps value imputation, discussed in chapter 6.
In the next chapter we turn to data formats that are hierarchically organized rather than tabular.